Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor - PubMed (original) (raw)
Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor
Heike Hofmann et al. Biochem Biophys Res Commun. 2004.
Abstract
The angiotensin converting enzyme 2 (ACE2) has been identified as a receptor for the severe acute respiratory syndrome associated coronavirus (SARS-CoV). Here we show that ACE2 expression on cell lines correlates with susceptibility to SARS-CoV S-driven infection, suggesting that ACE2 is a major receptor for SARS-CoV. The soluble ectodomain of ACE2 specifically abrogated S-mediated infection and might therefore be exploited for the generation of inhibitors. Deletion of a major portion of the cytoplasmic domain of ACE2 had no effect on S-driven infection, indicating that this domain is not important for receptor function. Our results point to a central role of ACE2 in SARS-CoV infection and suggest a minor contribution of the cytoplasmic domain to receptor function.
Figures
Fig. 1
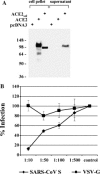
(A) Expression of ACE2 in cell culture cell lines susceptible (+) or refractory (−) to SARS-CoV infection. Total RNA was isolated from the indicated cell lines followed by reverse transcription. Subsequently, a nested PCR with ACE2-specific oligonucleotides was performed using either the resulting cDNAs as templates (middle panel, +RT) or employing the input RNA (upper panel, −RT). As a control, all cDNAs were subjected to a PCR with GAPDH-specific oligonucleotides (lower panel). (B) Enhanced SARS-CoV S-mediated entry into 293T cells transiently over-expressing ACE2. ACE2 of human (hu) and African green monkey (agm) origin or human CD13 were transiently expressed in 293T cells followed by infection with SARS-CoV S-pseudotypes carrying a luciferase reporter gene. After 72 h, cells were lysed and luciferase activity was determined in the cell extracts. Each experiment was performed in quadruplicate and repeated at least three times with independent virus stocks.
Fig. 2
Expression of soluble ACE2 protein and inhibition of SARS-CoV S-driven infection. (A) Expression of the soluble ACE2 ectodomain. Either a pcDNA3 control vector (lane 1), wild type ACE2 (lane 2) or an ACE2 variant comprising only the ectodomain (lane 3) was transiently expressed in 293T cells. After 48 h, cells and culture supernatants (lanes 4–6) were harvested and analyzed for ACE2 expression via Western blot. (B) Inhibition of S-mediated entry into 293T cells by soluble ACE2. S-bearing pseudotypes and VSV-G pseudotypes normalized for equal luciferase activity (104 c.p.s.) upon infection of target cells were pre-incubated with the indicated dilutions of concentrated soluble ACE2 and used for infection of 293T cells. Luciferase activity was determined in cell extracts after 72 h. The relative luciferase units obtained after infection in the absence of soluble ACE2 was set as 100%. Each experiment was performed in quadruplicate and repeated three times; similar results were obtained with a different soluble ACE2 preparation and with independent virus stocks.
Fig. 3
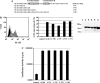
Analysis of the contribution of the ACE2 cytoplasmic domain to receptor function. (A) Schematic overview depicting the C-terminal ACE2 mutants analyzed. Putative tyrosine and casein kinase motifs are boxed. (B) Surface expression of ACE2 and the ACE2 deletion mutants. ACE2 was transiently expressed in 293T cells and analyzed by FACS using a polyclonal ACE2 antiserum followed by incubation with a polyclonal FITC-labeled secondary antibody (left panel, dark grey). As controls, pcDNA3-transfected cells were incubated with the secondary antibody (black line) or with both the ACE2-specific antiserum in combination with the secondary antibody (light grey). Similarly, the indicated ACE2 deletion mutants were subjected to FACS analysis; the percentage of ACE2 expressing cells is shown (middle panel). In parallel, expression of wild type ACE2 (lane 2) and all ACE2 mutants was examined by Western blot analysis (right panel: lane 1, pcDNA3; lane 3, mutant 1–790; lane 4, mutant 1–779; lane 5, mutant 1–775; and lane 6, mutant 1–771). (C) Role of the cytoplasmic domain within ACE2 for SARS-CoV S-mediated infection of target cells. ACE2 and the indicated deletion mutants were transiently expressed in 293T cells followed by infection with S-pseudotypes carrying luciferase as reporter gene. After 72 h, the luciferase activity was determined. Each experiment was performed in quadruplicate and repeated at least three times with independent virus preparations.
Similar articles
- ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia.
Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, Farzan M, Wohlford-Lenane C, Perlman S, McCray PB Jr. Jia HP, et al. J Virol. 2005 Dec;79(23):14614-21. doi: 10.1128/JVI.79.23.14614-14621.2005. J Virol. 2005. PMID: 16282461 Free PMC article. - Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus.
Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Li W, et al. Nature. 2003 Nov 27;426(6965):450-4. doi: 10.1038/nature02145. Nature. 2003. PMID: 14647384 Free PMC article. - Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2.
Moore MJ, Dorfman T, Li W, Wong SK, Li Y, Kuhn JH, Coderre J, Vasilieva N, Han Z, Greenough TC, Farzan M, Choe H. Moore MJ, et al. J Virol. 2004 Oct;78(19):10628-35. doi: 10.1128/JVI.78.19.10628-10635.2004. J Virol. 2004. PMID: 15367630 Free PMC article. - Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus.
Kuhn JH, Li W, Choe H, Farzan M. Kuhn JH, et al. Cell Mol Life Sci. 2004 Nov;61(21):2738-43. doi: 10.1007/s00018-004-4242-5. Cell Mol Life Sci. 2004. PMID: 15549175 Free PMC article. Review. - Receptor recognition and cross-species infections of SARS coronavirus.
Li F. Li F. Antiviral Res. 2013 Oct;100(1):246-54. doi: 10.1016/j.antiviral.2013.08.014. Epub 2013 Aug 29. Antiviral Res. 2013. PMID: 23994189 Free PMC article. Review.
Cited by
- Role and mechanism of angiotensin-converting enzyme 2 in acute lung injury in coronavirus disease 2019.
Liu MY, Zheng B, Zhang Y, Li JP. Liu MY, et al. Chronic Dis Transl Med. 2020 Jun;6(2):98-105. doi: 10.1016/j.cdtm.2020.05.003. Epub 2020 May 19. Chronic Dis Transl Med. 2020. PMID: 32550040 Free PMC article. Review. - [COVID-19, management, therapeutic and vaccine approaches].
Matusik É, Ayadi M, Picard N. Matusik É, et al. Actual Pharm. 2020 Oct;59(599):27-33. doi: 10.1016/j.actpha.2020.08.007. Epub 2020 Aug 21. Actual Pharm. 2020. PMID: 32863556 Free PMC article. French. - The emergence of COVID-19 as a global pandemic: Understanding the epidemiology, immune response and potential therapeutic targets of SARS-CoV-2.
Muralidar S, Ambi SV, Sekaran S, Krishnan UM. Muralidar S, et al. Biochimie. 2020 Dec;179:85-100. doi: 10.1016/j.biochi.2020.09.018. Epub 2020 Sep 22. Biochimie. 2020. PMID: 32971147 Free PMC article. Review. - Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes.
Temperton NJ, Chan PK, Simmons G, Zambon MC, Tedder RS, Takeuchi Y, Weiss RA. Temperton NJ, et al. Emerg Infect Dis. 2005 Mar;11(3):411-6. doi: 10.3201/eid1103.040906. Emerg Infect Dis. 2005. PMID: 15757556 Free PMC article. - Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations.
Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X, Wen F, Huang X, Ning G, Wang W. Cao Y, et al. Cell Discov. 2020 Feb 24;6:11. doi: 10.1038/s41421-020-0147-1. eCollection 2020. Cell Discov. 2020. PMID: 32133153 Free PMC article. No abstract available.
References
- Drosten C, Gunther S, Preiser W, van der W.S, Brodt H.R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier R.A, Berger A, Burguiere A.M, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra J.C, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk H.D, Osterhaus A.D, Schmitz H, Doerr H.W. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
- Ksiazek T.G, Erdman D, Goldsmith C.S, Zaki S.R, Peret T, Emery S, Tong S, Urbani C, Comer J.A, Lim W, Rollin P.E, Dowell S.F, Ling A.E, Humphrey C.D, Shieh W.J, Guarner J, Paddock C.D, Rota P, Fields B, DeRisi J, Yang J.Y, Cox N, Hughes J.M, LeDuc J.W, Bellini W.J, Anderson L.J. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
- Marra M.A, Jones S.J, Astell C.R, Holt R.A, Brooks-Wilson A, Butterfield Y.S, Khattra J, Asano J.K, Barber S.A, Chan S.Y, Cloutier A, Coughlin S.M, Freeman D, Girn N, Griffith O.L, Leach S.R, Mayo M, McDonald H, Montgomery S.B, Pandoh P.K, Petrescu A.S, Robertson A.G, Schein J.E, Siddiqui A, Smailus D.E, Stott J.M, Yang G.S, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth T.F, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples G.A, Tyler S, Vogrig R, Ward D, Watson B, Brunham R.C, Krajden M, Petric M, Skowronski D.M, Upton C, Roper R.L. Science. 2003;300:1399–1404. - PubMed
- Rota P.A, Oberste M.S, Monroe S.S, Nix W.A, Campagnoli R, Icenogle J.P, Penaranda S, Bankamp B, Maher K, Chen M.H, Tong S, Tamin A, Lowe L, Frace M, DeRisi J.L, Chen Q, Wang D, Erdman D.D, Peret T.C, Burns C, Ksiazek T.G, Rollin P.E, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus A.D, Drosten C, Pallansch M.A, Anderson L.J, Bellini W.J. Science. 2003;300:1394–1399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous