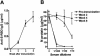An exposed domain in the severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies - PubMed (original) (raw)
. 2004 Jul;78(13):7217-26.
doi: 10.1128/JVI.78.13.7217-7226.2004.
Hong Wang, Danlin Luo, Thomas Rowe, Zheng Wang, Robert J Hogan, Shihong Qiu, Robert J Bunzel, Guoqiang Huang, Vinod Mishra, Thomas G Voss, Robert Kimberly, Ming Luo
Affiliations
- PMID: 15194798
- PMCID: PMC421657
- DOI: 10.1128/JVI.78.13.7217-7226.2004
An exposed domain in the severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies
Tong Zhou et al. J Virol. 2004 Jul.
Abstract
Exposed epitopes of the spike protein may be recognized by neutralizing antibodies against severe acute respiratory syndrome (SARS) coronavirus (CoV). A protein fragment (S-II) containing predicted epitopes of the spike protein was expressed in Escherichia coli. The properly refolded protein fragment specifically bound to the surface of Vero cells. Monoclonal antibodies raised against this fragment recognized the native spike protein of SARS CoV in both monomeric and trimeric forms. These monoclonal antibodies were capable of blocking S-II attachment to Vero cells and exhibited in vitro antiviral activity. These neutralizing antibodies mapped to epitopes in two peptides, each comprising 20 amino acids. Thus, this region of the spike protein might be a target for generation of therapeutic neutralizing antibodies against SARS CoV and for vaccine development to elicit protective humoral immunity.
Figures
FIG.1.
Antigenic analysis of the S protein. (A) Potential antigenic peptides predicted by EMBOSS:Antigenic. The scores are related to the probability that the amino acid sequence is an antigenic determinant on the basis of empirical data and the distribution of amino acids (9). (B) BLAST analysis of the S protein sequences of SARS CoV and HCoV 229E was performed with the full-length amino acid sequences of both proteins at the National Center for Biotechnology Information website. The letters between the two sequences are conserved residues, and the plus signs indicate homologous changes. The starting point corresponds to the first amino acid aligned by BLAST, and only the beginning portion of the alignment is shown here. The shaded region showed significant homology (60%) in the receptor binding region of the HCoV 229E S protein. (C) The codons for S-II were optimized for usage in E. coli, and 18 oligonucleotides grouped with sticky overlap ends were synthesized and annealed together as nicked double-stranded DNA and ligase treated. The synthesized cDNA was cloned into the expression vector pET100/D-TOPO with a His6 tag and an EK recognition site at the N terminus (Invitrogen), and protein expression was induced with IPTG in BL21/ED3 host cells. The inclusion bodies containing S-II were dissolved in 8 M urea, refolded, and purified by Ni affinity chromatography. The yield of purified, refolded, soluble S-II was >2 mg/liter. Lanes: 1, molecular mass marker; 2, denatured S-II expressed in E. coli; 3, purified, refolded S-II. (D) The purified, refolded S-II was biotinylated and used as a probe for specific binding to the surface of Vero cells. Vero E6 cells (106; American Type Culture Collection) were first incubated with a 20-μl volume of 10 μg of biotinylated S-II or a control His6-tagged S.Tag protein (Novagen) per ml at RT for 30 min and then further incubated with 20 μl of PE-conjugated streptavidin. The binding of S-II to Vero cells was demonstrated by flow cytometry analysis. S-II, solid; control, open. S-II showed selective binding to Vero cells compared to two recombinant protein fragments covering aa 145 to 480 (S-I) or 601 to 973 (S-III). S-RBD, S protein receptor binding domain. (E) To demonstrate dose-dependent binding of S-II to Vero cells, Vero cells were incubated with the indicated concentration of the S-II fragment (solid) or a control (open) and MFI was determined. (F) The same flow cytometry analysis as in panel D, using fragments S-I and S-III.
FIG.2.
Characterization of MAbs against S-II. (A) Vero cells either uninfected as controls or infected with 0.5 PFU of SARS CoV (Toronto-2 isolate) per ml per well for 24 h, WI38 cells infected with 229E, and B-SC-1 cells infected with OC43 for 72 h were cytospun onto slides, acetone fixed, and stained with 0.2 μg of HRP-conjugated MAbs (S34 and S78) per ml at RT for 40 min. The reaction was developed with DAB substrate. (B) To measure S protein in SARS CoV-infected Vero cells, whole-cell lysates were prepared from Vero cells infected with SARS CoV (Toronto-2 isolate). Numbers of viral PFU per milliliter of the supernatant from the cell lysates were determined at 6 × 105 PFU/ml by standard plaque assay on Vero E6 cells. Cell lysates were generated by mixing equal volumes of frozen cell lysate and 2× SDS sample buffer and boiling the mixture for 5 min. Cell lysates were dialyzed against PBS overnight prior to ELISA. ELISA plates were coated with 4 μg of each of the indicated MAbs per ml and blocked with 3% BSA. The cell lysate was diluted to the indicated concentrations and incubated at RT for 1 h. The captured S protein was detected with HRP-conjugated polyclonal goat anti-S antibody. (C) Western blot analysis of the S protein of SARS CoV. Whole-cell lysate from either SARS CoV-infected Vero cells obtained as described above (lane 1 and 2) or control noninfected Vero cells (lane 3 and 4) was separated by SDS-7% PAGE under both reducing (lane 1 and 3) and nonreducing (lane 2 and 4) conditions and blotted. The blots were probed with HRP-conjugated S26 or S78 at 4°C overnight and developed by chemiluminescence. (D) To immunoprecipitate SARS CoV, anti-S-II MAbs were conjugated to Sepharose 4B and incubated with 2 × 108 PFU of virus in a 1-ml total volume at RT for 30 min and then washed twice with PBS. The immunoprecipitated samples were dissolved in SDS denaturing buffer and separated by SDS-7% PAGE under nonreducing conditions. After transfer, the blot was probed with a mixture of HRP-conjugated anti-N and anti-S MAbs (lanes: 1, control; 2, S26; 3, S34; 4, S78; 5, S84). (E) Flow cytometry analysis of blocking activity of anti-S-II MAbs. Vero cells (106) were incubated with 10 μg of biotinylated S-II per ml in the presence of various concentrations of each MAb, followed by PE-conjugated streptavidin and determination of MFI by quantitative flow cytometry. (F) ELISA of blocking activity of anti-S-II MAbs. The membrane fraction of Vero E6 cells was lysed in 1% Triton-PBS and passed through an S-II-conjugated affinity column. The bound proteins were biotinylated on the column and eluted with pH 3.0 glycine-HCl buffer for the detecting agents. ELISA plate was coated with 5 μg of S-II per ml in PBS and incubated with 1 μg of biotinylated binding proteins of Vero cells per ml in the presence of various concentrations of anti-S-II MAbs. The plate-bound binding proteins were then detected with HRP-conjugated streptavidin. Maximum binding was determined as the OD values in the absence of MAb. IgG, immunoglobulin G.
FIG. 3.
Neutralizing activity of anti-S-II MAbs. Various concentrations of diluted MAbs were added to duplicate 24-well plates containing confluent monolayers of Vero76 cells in Iscove's modified Eagle medium-2% fetal bovine serum-2 mM
l
-glutamine-antibiotics. SARS CoV (Toronto-2 isolate) was added to each well at a final concentration of 0.5 PFU/ml, and plates were incubated for 72 h at 37°C in 5% CO2. Medium was removed from the wells, one set of plates was fixed with 80% acetone in PBS for immunostaining, and 0.1% SDS was added to the other set of plates for analysis by ELISA. (A) Quantitative measurement of the SARS CoV N protein for neutralizing activity. A quantitative ELISA of SARS CoV N protein was developed for measurement of viral infection (T.Z. et al., submitted). Briefly, an ELISA plate was coated with two MAbs against the N and C termini of the SARS CoV N protein. Whole-cell lysates (0.1% SDS) of infected cells were diluted 1:100 and 1:1,000 in 3% BSA-PBS and incubated in a MAb-coated plate. The captured N protein was then detected with HRP-conjugated polyclonal goat anti-N antibody. The N protein content was determined with the full-length of recombinant N protein as the standard. IgG, immunoglobulin G. (B) Western blot analysis of the N protein in MAb-treated Vero cells infected with SARS CoV. Five microliters of whole-cell lysate was separated by SDS-10% PAGE and blotted. The N protein was probed with an HRP-conjugated MAb against N protein. (C) Immunohistochemistry detection of SARS CoV-infected cells. Acetone-fixed cells were stained with an HRP-conjugated anti-N MAb and revealed by DBA substrate buffer. Photos were taken at a magnification of ×400. The values at the top are the concentrations of the MAbs used for neutralization. (D) Neutralizing epitope mapping of anti-S-II MAbs. A series of the truncated S-II peptides were expressed in E. coli with a His6 tag at the N terminus. The truncated peptides were used to coat ELISA plates, which were incubated with 1 μg of HRP-conjugated anti-S-II MAbs per ml. (E) Location of the neutralizing epitopes relative to the S-I, S-II, and S-III fragments within the S protein.
FIG. 3.
Neutralizing activity of anti-S-II MAbs. Various concentrations of diluted MAbs were added to duplicate 24-well plates containing confluent monolayers of Vero76 cells in Iscove's modified Eagle medium-2% fetal bovine serum-2 mM
l
-glutamine-antibiotics. SARS CoV (Toronto-2 isolate) was added to each well at a final concentration of 0.5 PFU/ml, and plates were incubated for 72 h at 37°C in 5% CO2. Medium was removed from the wells, one set of plates was fixed with 80% acetone in PBS for immunostaining, and 0.1% SDS was added to the other set of plates for analysis by ELISA. (A) Quantitative measurement of the SARS CoV N protein for neutralizing activity. A quantitative ELISA of SARS CoV N protein was developed for measurement of viral infection (T.Z. et al., submitted). Briefly, an ELISA plate was coated with two MAbs against the N and C termini of the SARS CoV N protein. Whole-cell lysates (0.1% SDS) of infected cells were diluted 1:100 and 1:1,000 in 3% BSA-PBS and incubated in a MAb-coated plate. The captured N protein was then detected with HRP-conjugated polyclonal goat anti-N antibody. The N protein content was determined with the full-length of recombinant N protein as the standard. IgG, immunoglobulin G. (B) Western blot analysis of the N protein in MAb-treated Vero cells infected with SARS CoV. Five microliters of whole-cell lysate was separated by SDS-10% PAGE and blotted. The N protein was probed with an HRP-conjugated MAb against N protein. (C) Immunohistochemistry detection of SARS CoV-infected cells. Acetone-fixed cells were stained with an HRP-conjugated anti-N MAb and revealed by DBA substrate buffer. Photos were taken at a magnification of ×400. The values at the top are the concentrations of the MAbs used for neutralization. (D) Neutralizing epitope mapping of anti-S-II MAbs. A series of the truncated S-II peptides were expressed in E. coli with a His6 tag at the N terminus. The truncated peptides were used to coat ELISA plates, which were incubated with 1 μg of HRP-conjugated anti-S-II MAbs per ml. (E) Location of the neutralizing epitopes relative to the S-I, S-II, and S-III fragments within the S protein.
FIG. 4.
S-II elicits a protective antibody response against SARS CoV. Six- to 8-week-old female BALB/c mice were subcutaneously immunized with 100 μg of S-II in Freund's complete adjuvant and boosted twice with S-II in PBS weekly. Serum samples were collected at the indicated time points after primary immunization. (A) Serum levels of anti-S-II antibody were measured by indirect ELISA with an anti-S-II MAb as the standard. IgG, immunoglobulin G. (B) The 10-fold-diluted sera were added to Vero E6 cell cultures 1 h prior to infection of each well with 0.5 PFU of SARS CoV per ml. The neutralizing activity of antisera was determined by quantitative measurement of N protein by ELISA as described above. The results are presented as the mean ± the standard deviation of two pools of six mice at each time point.
Similar articles
- Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acute respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants.
Sui J, Li W, Roberts A, Matthews LJ, Murakami A, Vogel L, Wong SK, Subbarao K, Farzan M, Marasco WA. Sui J, et al. J Virol. 2005 May;79(10):5900-6. doi: 10.1128/JVI.79.10.5900-5906.2005. J Virol. 2005. PMID: 15857975 Free PMC article. - Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies.
Zhang H, Wang G, Li J, Nie Y, Shi X, Lian G, Wang W, Yin X, Zhao Y, Qu X, Ding M, Deng H. Zhang H, et al. J Virol. 2004 Jul;78(13):6938-45. doi: 10.1128/JVI.78.13.6938-6945.2004. J Virol. 2004. PMID: 15194770 Free PMC article. - Vaccine design for severe acute respiratory syndrome coronavirus.
He Y, Jiang S. He Y, et al. Viral Immunol. 2005;18(2):327-32. doi: 10.1089/vim.2005.18.327. Viral Immunol. 2005. PMID: 16035944 Review. - SARS vaccine development.
Jiang S, He Y, Liu S. Jiang S, et al. Emerg Infect Dis. 2005 Jul;11(7):1016-20. doi: 10.3201/1107.050219. Emerg Infect Dis. 2005. PMID: 16022774 Free PMC article. Review.
Cited by
- Adenoviral expression of a truncated S1 subunit of SARS-CoV spike protein results in specific humoral immune responses against SARS-CoV in rats.
Liu RY, Wu LZ, Huang BJ, Huang JL, Zhang YL, Ke ML, Wang JM, Tan WP, Zhang RH, Chen HK, Zeng YX, Huang W. Liu RY, et al. Virus Res. 2005 Sep;112(1-2):24-31. doi: 10.1016/j.virusres.2005.02.009. Virus Res. 2005. PMID: 16022898 Free PMC article. - Identification and characterization of a neutralizing-epitope-containing spike protein fragment in turkey coronavirus.
Chen YN, Wu CC, Lin TL. Chen YN, et al. Arch Virol. 2011 Sep;156(9):1525-35. doi: 10.1007/s00705-011-1020-1. Epub 2011 May 19. Arch Virol. 2011. PMID: 21594597 Free PMC article. - Template-based coiled-coil antigens elicit neutralizing antibodies to the SARS-coronavirus.
Tripet B, Kao DJ, Jeffers SA, Holmes KV, Hodges RS. Tripet B, et al. J Struct Biol. 2006 Aug;155(2):176-94. doi: 10.1016/j.jsb.2006.03.019. Epub 2006 Apr 27. J Struct Biol. 2006. PMID: 16697221 Free PMC article. - Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region.
Chen Z, Zhang L, Qin C, Ba L, Yi CE, Zhang F, Wei Q, He T, Yu W, Yu J, Gao H, Tu X, Gettie A, Farzan M, Yuen KY, Ho DD. Chen Z, et al. J Virol. 2005 Mar;79(5):2678-88. doi: 10.1128/JVI.79.5.2678-2688.2005. J Virol. 2005. PMID: 15708987 Free PMC article. - Cell adhesion as a novel approach to determining the cellular binding motif on the severe acute respiratory syndrome coronavirus spike protein.
Chang HH, Chen PK, Lin GL, Wang CJ, Liao CH, Hsiao YC, Dong JH, Sun DS. Chang HH, et al. J Virol Methods. 2014 Jun;201:1-6. doi: 10.1016/j.jviromet.2014.01.022. Epub 2014 Feb 13. J Virol Methods. 2014. PMID: 24530430 Free PMC article.
References
- Dveksler, G. S., C. W. Dieffenbach, C. B. Cardellichio, K. McCuaig, M. N. Pensiero, G.-S. Jiang, N. Beauchemin, and K. V. Holmes. 1993. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus-A59. J. Virol. 67:1-8. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous