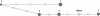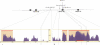Identifying candidate causal variants responsible for altered activity of the ABCB1 multidrug resistance gene - PubMed (original) (raw)
. 2004 Jul;14(7):1333-44.
doi: 10.1101/gr.1965304. Epub 2004 Jun 14.
Affiliations
- PMID: 15197162
- PMCID: PMC442149
- DOI: 10.1101/gr.1965304
Identifying candidate causal variants responsible for altered activity of the ABCB1 multidrug resistance gene
Nicole Soranzo et al. Genome Res. 2004 Jul.
Abstract
The difficulty of fine localizing the polymorphisms responsible for genotype-phenotype correlations is emerging as an important constraint in the implementation and interpretation of genetic association studies, and calls for the definition of protocols for the follow-up of associated variants. One recent example is the 3435C>T polymorphism in the multidrug transporter gene ABCB1, associated with protein expression and activity, and with several clinical conditions. Available data suggest that 3435C>T may not directly cause altered transport activity, but may be associated with one or more causal variants in the poorly characterized stretch of linkage disequilibrium (LD) surrounding it. Here we describe a strategy for the follow-up of reported associations, including a Bayesian formalization of the associated interval concept previously described by Goldstein. We focus on the region of high LD around 3435C>T to compile an exhaustive list of variants by (1) using a relatively coarse set of marker typings to assess the pattern of LD, and (2) resequencing derived and ancestral chromosomes at 3435C>T through the associated interval. We identified three intronic sites that are strongly associated with the 3435C>T polymorphism. One of them is associated with multidrug resistance in patients with epilepsy (chi2 = 3.78, P = 0.052), and sits within a stretch of significant evolutionary conservation. We argue that these variants represent additional candidates for influencing multidrug resistance due to P-glycoprotein activity, with the IVS 26+80 T>C being the best candidate among the three intronic sites. Finally, we describe a set of six haplotype tagging single-nucleotide polymorphisms that represent common ABCB1 variation surrounding 3435C>T in Europeans.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures
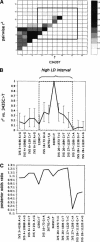
Figure 1
(A) Graph of pairwise r2 measures of LD among the 14 high-frequency (>6%) ABCB1 SNP loci described in Table 1. (B) r2 for 3435C>T against each of the remainder SNPs, together with Bayesian 95% credible intervals calculated by the method described in the Supplemental materials. The dashed box identifies the high-LD interval around 3435C>T. All sites upstream of IVS 27+1220 show association with 3435C>T, significant at the 0.001 level (Fisher's exact test). At the loci downstream of this site, we observe no significant association (P > 0.05) with 3435C>T. Throughout the study we used r2 to measure association, to take into account the dependency of LD on allelic frequencies. (C) Graph of posterior odds of each SNP being causal relative to 3435C>T being causal, based on the combined LD data and case control data on 3435C>T (Siddiqui et al. 2003), using the method described in the Supplemental materials.
Figure 2
Reduced-median haplotype network of ABCB1 haplotypes found with frequencies >5% in the set of 24 CEPH trios, as described in Table 2. Loci include IVS 6+139C>T, IVS 6+251A>G, 1236C>T, IVS 16+73G>A, 2677G>T,A, 3435C>T, IVS 26+1573T>C, and IVS 26+1684 of Table 1. Node size is proportional to haplotype frequency. Mutated positions are indicated by white dots. The ancestral haplotype was determined by resequencing of a chimpanzee sample (gray).
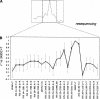
Figure 3
The high-LD interval (A, dashed box) was fully sequenced in two chromosomes, leading to the identification of candidate causal sites; resequencing of these variants in CEPH trios was used to resolve LD structure within this interval (B). Of all homozygous variants, those that are in high or complete r2 with 3435C>T (•) are the most likely candidates to be causal; heterozygous mutations (▵) result in decreased r2. (○) The cases where a new set of CEPH trios was used to calculate r2 compared to the rest of the study. Bars indicate the Bayesian 95% credible intervals calculated by the method described in Supplemental materials.
Figure 4
Position of the three candidate causal mutations within ABCB1, showing the position of the three new candidate variants in high LD with 3435C>T. In the boxes are the two regions further investigated by phylogenetic shadowing. Plots of cumulative nucleotide divergence were generated using the eShadow tool by amplifying the region surrounding IVS 25+3050 G>T (A) and 3435C>T (B) in 17 and 15 primate species, respectively. Orange shading indicates the region of significant conservation; the horizontal red bar at nucleotides 715–921 of the alignment in B indicates the position of exon 26. The position of the three intronic sites is marked by a vertical dashed line: IVS 25+3050 G>T at nucleotide 710 (A), and IVS 25+5231 T>C, 3435C>T, and IVS 26+80 T>C sites at 184, 867, and 1001 bp of the alignment (B).
Figure 5
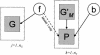
Description of Bayesian models for assessment of the associated interval (see text). The link between f and P is dashed to indicate that it is present in Model 1 but not Model 0.
Similar articles
- Association of ABCB1 genetic variants 3435C>T and 2677G>T to ABCB1 mRNA and protein expression in brain tissue from refractory epilepsy patients.
Mosyagin I, Runge U, Schroeder HW, Dazert E, Vogelgesang S, Siegmund W, Warzok RW, Cascorbi I. Mosyagin I, et al. Epilepsia. 2008 Sep;49(9):1555-61. doi: 10.1111/j.1528-1167.2008.01661.x. Epub 2008 May 20. Epilepsia. 2008. PMID: 18494787 - Predisposition to epilepsy--does the ABCB1 gene play a role?
Nurmohamed L, Garcia-Bournissen F, Buono RJ, Shannon MW, Finkelstein Y. Nurmohamed L, et al. Epilepsia. 2010 Sep;51(9):1882-5. doi: 10.1111/j.1528-1167.2010.02588.x. Epilepsia. 2010. PMID: 20491876 Review. - Promoter polymorphisms and allelic imbalance in ABCB1 expression.
Loeuillet C, Weale M, Deutsch S, Rotger M, Soranzo N, Wyniger J, Lettre G, Dupré Y, Thuillard D, Beckmann JS, Antonarakis SE, Goldstein DB, Telenti A. Loeuillet C, et al. Pharmacogenet Genomics. 2007 Nov;17(11):951-9. doi: 10.1097/FPC.0b013e3282eff934. Pharmacogenet Genomics. 2007. PMID: 18075465 - No association of ABCB1 polymorphisms with drug-refractory epilepsy in a north Indian population.
Lakhan R, Misra UK, Kalita J, Pradhan S, Gogtay NJ, Singh MK, Mittal B. Lakhan R, et al. Epilepsy Behav. 2009 Jan;14(1):78-82. doi: 10.1016/j.yebeh.2008.08.019. Epub 2008 Oct 8. Epilepsy Behav. 2009. PMID: 18812236 - On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci.
Garner C, Slatkin M. Garner C, et al. Genet Epidemiol. 2003 Jan;24(1):57-67. doi: 10.1002/gepi.10217. Genet Epidemiol. 2003. PMID: 12508256 Review.
Cited by
- Effects of ABCB1 gene polymorphisms on autonomic nervous system activity during atypical antipsychotic treatment in schizophrenia.
Hattori S, Suda A, Kishida I, Miyauchi M, Shiraishi Y, Fujibayashi M, Tsujita N, Ishii C, Ishii N, Moritani T, Taguri M, Hirayasu Y. Hattori S, et al. BMC Psychiatry. 2018 Jul 17;18(1):231. doi: 10.1186/s12888-018-1817-5. BMC Psychiatry. 2018. PMID: 30016952 Free PMC article. - Nucleic Acid Aptamers for Molecular Therapy of Epilepsy and Blood-Brain Barrier Damages.
Zamay TN, Zamay GS, Shnayder NA, Dmitrenko DV, Zamay SS, Yushchenko V, Kolovskaya OS, Susevski V, Berezovski MV, Kichkailo AS. Zamay TN, et al. Mol Ther Nucleic Acids. 2020 Mar 6;19:157-167. doi: 10.1016/j.omtn.2019.10.042. Epub 2019 Nov 15. Mol Ther Nucleic Acids. 2020. PMID: 31837605 Free PMC article. Review. - Etiologies of Multidrug-Resistant Epilepsy in Latin America: A Comprehensive Review of Structural, Genetic, Metabolic, Inflammatory, and Infectious Origins: A Systematic Review.
Hinojosa-Figueroa MS, Cruz-Caraguay M, Torres Pasquel A, Puga Rosero V, Eguiguren Chavez CB, Rodas JA, Leon-Rojas JE. Hinojosa-Figueroa MS, et al. Biomolecules. 2025 Apr 12;15(4):576. doi: 10.3390/biom15040576. Biomolecules. 2025. PMID: 40305305 Free PMC article. - Linkage and association analysis of CACNG3 in childhood absence epilepsy.
Everett KV, Chioza B, Aicardi J, Aschauer H, Brouwer O, Callenbach P, Covanis A, Dulac O, Eeg-Olofsson O, Feucht M, Friis M, Goutieres F, Guerrini R, Heils A, Kjeldsen M, Lehesjoki AE, Makoff A, Nabbout R, Olsson I, Sander T, Sirén A, McKeigue P, Robinson R, Taske N, Rees M, Gardiner M. Everett KV, et al. Eur J Hum Genet. 2007 Apr;15(4):463-72. doi: 10.1038/sj.ejhg.5201783. Epub 2007 Jan 31. Eur J Hum Genet. 2007. PMID: 17264864 Free PMC article. - Associations between the C3435T polymorphism of the ABCB1 gene and drug resistance in epilepsy: a meta-analysis.
Lv WP, Han RF, Shu ZR. Lv WP, et al. Int J Clin Exp Med. 2014 Nov 15;7(11):3924-32. eCollection 2014. Int J Clin Exp Med. 2014. PMID: 25550900 Free PMC article.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
- Bellamy, W.T. 1996. P-glycoproteins and multidrug resistance. Annu. Rev. Pharmacol. Toxicol. 36: 161–183. - PubMed
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391–1394. - PubMed
- Cornwell, M.M. and Smith, D.E. 1993. SP1 activates the MDR1 promoter through one of two distinct G-rich regions that modulate promoter activity. J. Biol. Chem. 268: 19505–19511. - PubMed
WEB SITE REFERENCES
- http://popgen.biol.ucl.ac.uk/supdata.html; primer information.
- http://popgen.biol.ucl.ac.uk/software.html; TagIT and routines for the Bayesian analysis of the associated interval.
- http://www.cbil.upenn.edu/tess; TESS algorithm.
- http://eshadow.dcode.org/; eShadow.
- http://web1.iop.kcl.ac.uk/IoP/Departments/PsychMed/GEpiBSt/software.shtml; EH and PM programs.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials