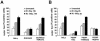Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin - PubMed (original) (raw)
Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin
XiaoZhe Wang et al. Mol Cell Biol. 2004 Jul.
Abstract
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility syndrome with at least 11 complementation groups (A, B, C, D1, D2, E, F, G, I, J, and L), and eight FA genes have been cloned. The FANCD1 gene is identical to the breast cancer susceptibility gene, BRCA2. The FA proteins cooperate in a common pathway, but the function of BRCA2/FANCD1 in this pathway remains unknown. Here we show that monoubiquitination of FANCD2, which is activated by DNA damage, is required for targeting of FANCD2 to chromatin, where it interacts with BRCA2. FANCD2-Ub then promotes BRCA2 loading into a chromatin complex. FANCD2(-/-) cells are deficient in the assembly of DNA damage-inducible BRCA2 foci and in chromatin loading of BRCA2. Functional complementation with the FANCD2 cDNA restores BRCA2 foci and its chromatin loading following DNA damage. BRCA2(-/-) cells expressing a carboxy-terminal truncated BRCA2 protein form IR-inducible BRCA2 and FANCD2 foci, but these foci fail to colocalize. Functional complementation of these cells with wild-type BRCA2 restores the interaction of BRCA2 and FANCD2. The C terminus of BRCA2 is therefore required for the functional interaction of BRCA2 and FANCD2 in chromatin. Taken together, our results demonstrate that monoubiquitination of FANCD2, which is regulated by the FA pathway, promotes BRCA2 loading into chromatin complexes. These complexes appear to be required for normal homology-directed DNA repair.
Figures
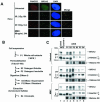
FIG. 1.
Colocalization, cofractionation, and coimmunoprecipitation of monoubiquitinated FANCD2 and BRCA2 in chromatin. (A) Colocalization of FANCD2 and BRCA2 in DNA damage-inducible foci. HeLa cells were either untreated, exposed to IR (2 or 15 Gy), or treated with MMC (80 ng/ml), as indicated. HeLa cells were double stained with polyclonal anti-FANCD2 (E35) (red) and monoclonal anti-BRCA2 (Ab-1) (green) antibodies after the indicated time of treatment. Magnification, ×630. (B) Protocol utilized for nuclear fractionation of cells. Cytoplasm and nucleoplasm were extracted by permeabilization with detergent, the resulting nuclei were DNase I digested, and chromatin was extracted with ammonium sulfate (NH2SO4). (C) U2OS cells, either untreated or exposed to IR (15 Gy) or MMC (170 ng/ml), were fractionated 15 h after the initiation of DNA damage. Supernatants (S) and pellets (P) were subjected to Western blot analysis with the indicated antibodies: E35 for FANCD2 and Ab-1 for BRCA2. The presence of chromatin in the S4 fraction was confirmed by blotting with anti-histone H4 antibody. (D) S2 (soluble nuclear proteins) and S4 (chromatin) fractions of U2OS cells were subjected to immunoprecipitation with mouse monoclonal anti-BRCA2 antibody (Ab-1) or a control mouse antibody (mIgG) and then immunoblotted with either polyclonal anti-BRCA2 (Ab-2) or anti-FANCD2 (E35) antibodies. Heavy chain IgG was used as a loading control. (E) HeLa cells were transfected with a cDNA encoding HA-ubiquitin, as indicated. After transfection, cells were treated with the indicated dose of IR or MMC. S2 (soluble nuclear proteins) and S4 (chromatin) fractions of HeLa cells were immunoprecipitated (IP) with a polyclonal antibody (E35) to FANCD2, as indicated. Immune complexes were run on SDS-PAGE and immunoblotted with anti-FANCD2 (FI-17) or anti-HA (HA.11) monoclonal antibodies. WCE, whole-cell extract.
FIG. 1.
Colocalization, cofractionation, and coimmunoprecipitation of monoubiquitinated FANCD2 and BRCA2 in chromatin. (A) Colocalization of FANCD2 and BRCA2 in DNA damage-inducible foci. HeLa cells were either untreated, exposed to IR (2 or 15 Gy), or treated with MMC (80 ng/ml), as indicated. HeLa cells were double stained with polyclonal anti-FANCD2 (E35) (red) and monoclonal anti-BRCA2 (Ab-1) (green) antibodies after the indicated time of treatment. Magnification, ×630. (B) Protocol utilized for nuclear fractionation of cells. Cytoplasm and nucleoplasm were extracted by permeabilization with detergent, the resulting nuclei were DNase I digested, and chromatin was extracted with ammonium sulfate (NH2SO4). (C) U2OS cells, either untreated or exposed to IR (15 Gy) or MMC (170 ng/ml), were fractionated 15 h after the initiation of DNA damage. Supernatants (S) and pellets (P) were subjected to Western blot analysis with the indicated antibodies: E35 for FANCD2 and Ab-1 for BRCA2. The presence of chromatin in the S4 fraction was confirmed by blotting with anti-histone H4 antibody. (D) S2 (soluble nuclear proteins) and S4 (chromatin) fractions of U2OS cells were subjected to immunoprecipitation with mouse monoclonal anti-BRCA2 antibody (Ab-1) or a control mouse antibody (mIgG) and then immunoblotted with either polyclonal anti-BRCA2 (Ab-2) or anti-FANCD2 (E35) antibodies. Heavy chain IgG was used as a loading control. (E) HeLa cells were transfected with a cDNA encoding HA-ubiquitin, as indicated. After transfection, cells were treated with the indicated dose of IR or MMC. S2 (soluble nuclear proteins) and S4 (chromatin) fractions of HeLa cells were immunoprecipitated (IP) with a polyclonal antibody (E35) to FANCD2, as indicated. Immune complexes were run on SDS-PAGE and immunoblotted with anti-FANCD2 (FI-17) or anti-HA (HA.11) monoclonal antibodies. WCE, whole-cell extract.
FIG. 2.
Monoubiquitinated FANCD2 promotes IR-induced assembly of BRCA2 foci. (A) Whole-cell extracts of PD20 (FA-D2) fibroblasts stably expressing empty vector (PD20F + Vec) and PD20 fibroblasts corrected with FANCD2 (PD20F + FANCD2) were subjected to immunoprecipitation with mouse monoclonal anti-BRCA2 (Ab-1) or a control mouse antibody (mIgG) and then immunoblotted with either anti-FANCD2 (E35) or anti-BRCA2 (Ab-2) antibodies. Heavy chain IgG was used as a loading control. (B) Formation of subnuclear FANCD2 and BRCA2 foci in response to IR treatment. PD20 (FA-D2) fibroblasts, stably expressing empty vector alone (upper panel) or full-length FANCD2 cDNA (lower panel), were either untreated or treated with IR (15 Gy) and fixed 4 h later. Cells were double stained with polyclonal anti-FANCD2 (E35) (red) and monoclonal anti-BRCA2 (Ab-1) (green) antibodies and analyzed by immunofluorescence microscopy. Magnification, ×630. (C) Quantification of BRCA2 foci. PD20 (FA-D2) fibroblasts stably expressing empty vector alone (PD20F + Vec), FANCD2 (PD20F + FANCD2), or FANCD2-K561R mutant (PD20F + K561R), GM6914 (FA-A) fibroblasts and corrected GM6914 fibroblasts stably expressing FANCA (GM6914 + FANCA), EUFA130 (FA-E) lymphoblasts and corrected EUFA130 lymphoblasts (EUFA130 + HA-FANCE), and HeLa cells were either untreated or treated with IR (15 Gy) and fixed 4 h later. Cells with more than four distinct foci were counted as positive. A total of 200 cells/sample were analyzed. The values shown are the mean ± standard deviation from three separate experiments.
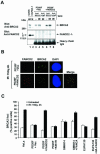
FIG. 3.
FANCD2 and BRCA2 foci fail to associate in FA-D1 (BRCA2−/−) cells. (A) Schematic diagram of human BRCA2 protein, indicating mutations in EUFA 423 (FA-D1) cells. The positions of epitopes for the anti-BRCA2 antibodies Ab-1 and Ab-2 are shown. (B) EUFA423 (FA-D1) fibroblasts and corrected cells stably transfected with human chromosome 13 (EUFA423 + BRCA2) were exposed to IR, either at different doses (left) or for different periods of time (right). Western blotting was performed with anti-BRCA2 (Ab-1 or Ab-2) or anti-FANCD2 (E35) antibodies. (C) Immunofluorescent localization of BRCA2 and FANCD2 following treatment with IR (15 Gy) was examined in EUFA423 (FA-D1) fibroblasts, EUFA423 fibroblasts transiently transfected with pcDNA3 HA-BRCA2 (EUFA423 + HA-BRCA2), and EUFA 423 fibroblasts stably transfected with human chromosome 13 (EUFA423 + BRCA2). Cells were double stained with the indicated antibodies and analyzed by immunofluorescence microscopy. Magnification, ×630. (D and E) Quantification of the percentage of cells with FANCD2 foci (D) and the percentage of cells with colocalization of FANCD2 and BRCA2 foci (E) in HeLa cells, EUFA423 fibroblasts, EUFA423 fibroblasts transiently expressing HA-BRCA2 (EUFA423 + HA-BRCA2), and EUFA423 fibroblasts stably expressing human chromosome 13 (EUFA423 + BRCA2). Cells were either untreated or treated with IR (15 Gy) and fixed 4 h later for immunofluorescence microscopy. Cells with more than four distinct foci were counted as positive. Two hundred and 100 cells/sample were analyzed in panels D and E, respectively. The values shown are the mean ± standard deviation from three separate experiments. (F) S2 (soluble nuclear proteins) and S4 (chromatin) fractions of irradiated EUFA423 cells and EUFA423 cells stably expressing human chromosome 13 (EUFA423 + BRCA2) were subjected to immunoprecipitation with rabbit polyclonal anti-FANCD2 antibody (E35) or a control nonimmunized rabbit serum (Pre-imm) and then immunoblotted with either monoclonal anti-BRCA2 (Ab-1) or anti-FANCD2 (FI-17) antibodies. Heavy chain IgG was used as a loading control. WCE, whole-cell extract.
FIG. 3.
FANCD2 and BRCA2 foci fail to associate in FA-D1 (BRCA2−/−) cells. (A) Schematic diagram of human BRCA2 protein, indicating mutations in EUFA 423 (FA-D1) cells. The positions of epitopes for the anti-BRCA2 antibodies Ab-1 and Ab-2 are shown. (B) EUFA423 (FA-D1) fibroblasts and corrected cells stably transfected with human chromosome 13 (EUFA423 + BRCA2) were exposed to IR, either at different doses (left) or for different periods of time (right). Western blotting was performed with anti-BRCA2 (Ab-1 or Ab-2) or anti-FANCD2 (E35) antibodies. (C) Immunofluorescent localization of BRCA2 and FANCD2 following treatment with IR (15 Gy) was examined in EUFA423 (FA-D1) fibroblasts, EUFA423 fibroblasts transiently transfected with pcDNA3 HA-BRCA2 (EUFA423 + HA-BRCA2), and EUFA 423 fibroblasts stably transfected with human chromosome 13 (EUFA423 + BRCA2). Cells were double stained with the indicated antibodies and analyzed by immunofluorescence microscopy. Magnification, ×630. (D and E) Quantification of the percentage of cells with FANCD2 foci (D) and the percentage of cells with colocalization of FANCD2 and BRCA2 foci (E) in HeLa cells, EUFA423 fibroblasts, EUFA423 fibroblasts transiently expressing HA-BRCA2 (EUFA423 + HA-BRCA2), and EUFA423 fibroblasts stably expressing human chromosome 13 (EUFA423 + BRCA2). Cells were either untreated or treated with IR (15 Gy) and fixed 4 h later for immunofluorescence microscopy. Cells with more than four distinct foci were counted as positive. Two hundred and 100 cells/sample were analyzed in panels D and E, respectively. The values shown are the mean ± standard deviation from three separate experiments. (F) S2 (soluble nuclear proteins) and S4 (chromatin) fractions of irradiated EUFA423 cells and EUFA423 cells stably expressing human chromosome 13 (EUFA423 + BRCA2) were subjected to immunoprecipitation with rabbit polyclonal anti-FANCD2 antibody (E35) or a control nonimmunized rabbit serum (Pre-imm) and then immunoblotted with either monoclonal anti-BRCA2 (Ab-1) or anti-FANCD2 (FI-17) antibodies. Heavy chain IgG was used as a loading control. WCE, whole-cell extract.
FIG. 4.
FA-D1 and FA-D2 cells are defective in the assembly of IR-inducible RAD51 foci. (A) Quantification of RAD51 foci in EUFA423 (FA-D1) fibroblasts, EUFA423 fibroblasts stably transfected with human chromosome 13 (EUFA423 + BRCA2), and HeLa cells. Cells were either untreated or treated with IR (2 or 15 Gy) and fixed 15 h later. Cells were stained with monoclonal anti-RAD51 antibody and analyzed by immunofluorescence microscopy. Cells with more than four distinct foci were counted as positive. Two hundred cells/sample were analyzed. The values shown are the mean ± standard deviation from three separate experiments. (B) Quantification of RAD51 foci in PD20 (FA-D2) fibroblasts stably expressing empty vector alone (PD20F + Vec), full-length FANCD2 cDNA (PD20F + FANCD2), or the FANCD2 K561R mutant (PD20F + K561R), and HeLa cells. Cells were either untreated or treated with IR (2 or 15 Gy) and fixed 8 h later. Immunofluorescence microscopy and counts were performed as described above for panel A.
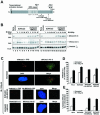
FIG. 5.
Monoubiquitination of FANCD2 promotes IR-inducible accumulation of BRCA2 in chromatin. (A) S2 (soluble nuclear proteins) and S4 (chromatin) fractions were prepared from U2OS and FA cells, which were either untreated or treated with IR (15 Gy, fractionated after 15 h). Fractions were subjected to Western blot analysis with anti-FANCD2 (E35) or anti-BRCA2 (Ab-1) antibodies. The extraction of chromatin in the S4 fraction was confirmed by blotting with anti-histone H4 antibody, as indicated. The FA cells are represented by GM6914 (FA-A) fibroblasts, EUFA423 (FA-D1) fibroblasts, and HSC230 (FA-B) lymphoblasts. (B) S2 (soluble nuclear proteins) and S4 (chromatin) fractions were prepared from PD20 (FA-D2) fibroblasts stably expressing empty vector alone (PD20F + HA-PMMP), HA-FANCD2 (PD20F + HA-FANCD2), or FANCD2-HA-K561R mutant (PD20F + HA-K561R), which were either untreated or treated with IR (15 Gy, fractionated after 6 h). Fractions were subjected to Western blot analysis with anti-FANCD2 (E35) or anti-BRCA2 (Ab-1) antibodies. The extraction of chromatin in the S4 fraction was confirmed by blotting with anti-histone H4 antibody.
FIG. 6.
ATM is required for IR-activated phosphorylation of BRCA2 in chromatin. (A) BRCA2 immunoprecipitated (Ab-1) from chromatin fractions was treated either with or without λ-phosphatase and phosphatase inhibitors, as indicated. Chromatin fractions were derived either from untreated U2OS cells or 15 h following IR treatment (15 Gy). The mobility shift of BRCA2 is shown in lanes 4 and 6. WCE, whole-cell extract. (B) BRCA2 derived from the chromatin fraction of corrected AT (AT + ATM) fibroblasts at time points after treatment with IR (15 Gy) undergoes a mobility shift, while BRCA2 from the chromatin fraction of AT (ATM−/−) fibroblasts does not. Chromatin fractions were immunoblotted with either anti-BRCA2 (Ab-1), anti-ATM, or anti-FANCD2 (E35) antibodies. (C) EUFA423 (FA-D1) and PD20 (FA-D2) cells are defective in an IR-inducible S-phase checkpoint. RDS was assessed 30 min after delivery of IR to the indicated cell lines.
FIG. 7.
Interaction of FANCE and BRCA2. (A) Whole-cell extracts (WCE) from HeLa cells, either untreated or exposed to IR (15 Gy, harvested 15 h later) or MMC (40 ng/ml, harvested 24 h later), were subjected to immunoprecipitation with mouse monoclonal anti-BRCA2 antibody (Ab-1) or a control mouse antibody (mIgG). Heavy chain IgG was used as a loading control. (B) Whole-cell extracts from unirradiated or irradiated EUFA130 (FA-E), and EUFA130 cells stably expressing HA-FANCE (EUFA130 + HA-FANCE), were subjected to immunoprecipitation with polyclonal anti-BRCA2 antibody (H-300) or a control rabbit antibody (rIgG) and then immunoblotted with anti-BRCA2 (Ab-1) or anti-HA antibodies. Heavy chain IgG was used as a loading control. (C) EUFA130 (FA-E) lymphoblasts and EUFA130 cells stably expressing HA-FANCE (EUFA130 + HA-FANCE) were either untreated or exposed to IR at different doses, as indicated, and harvested after 4 h. Western blotting was performed with anti-BRCA2 (Ab-1), anti-FANCD2 (E35), or anti-HA antibodies. (D) Reciprocal coimmunoprecipitation of FANCD2 and BRCA2 with other antibodies. The indicated fractions (S2 and S4), prepared from irradiated U2OS cells, were immunoprecipitated with antibody to FANCD2 (E35), FANCE, or a control nonimmunized rabbit serum (Pre-imm), and the immune complexes were immunoblotted with anti-BRCA2 (Ab-1) or anti-FANCD2 (FI-17) antibodies. Heavy chain IgG was used as a loading control. (E) Schematic model of the ATM-BRCA2-FANCD2-mediated DNA damage response. IR activates ATM, resulting in the phosphorylation of BRCA2, FANCD2, and several other protein substrates. Activated BRCA2 is then recruited to chromatin by FANCD2, which is activated by monoubiquitination. The interaction of BRCA2 and FANCD2 requires both the C terminus of BRCA2 and FANCD2 monoubiquitination. Following its loading onto chromatin, BRCA2 then functions downstream of monubiquitinated FANCD2.
FIG. 7.
Interaction of FANCE and BRCA2. (A) Whole-cell extracts (WCE) from HeLa cells, either untreated or exposed to IR (15 Gy, harvested 15 h later) or MMC (40 ng/ml, harvested 24 h later), were subjected to immunoprecipitation with mouse monoclonal anti-BRCA2 antibody (Ab-1) or a control mouse antibody (mIgG). Heavy chain IgG was used as a loading control. (B) Whole-cell extracts from unirradiated or irradiated EUFA130 (FA-E), and EUFA130 cells stably expressing HA-FANCE (EUFA130 + HA-FANCE), were subjected to immunoprecipitation with polyclonal anti-BRCA2 antibody (H-300) or a control rabbit antibody (rIgG) and then immunoblotted with anti-BRCA2 (Ab-1) or anti-HA antibodies. Heavy chain IgG was used as a loading control. (C) EUFA130 (FA-E) lymphoblasts and EUFA130 cells stably expressing HA-FANCE (EUFA130 + HA-FANCE) were either untreated or exposed to IR at different doses, as indicated, and harvested after 4 h. Western blotting was performed with anti-BRCA2 (Ab-1), anti-FANCD2 (E35), or anti-HA antibodies. (D) Reciprocal coimmunoprecipitation of FANCD2 and BRCA2 with other antibodies. The indicated fractions (S2 and S4), prepared from irradiated U2OS cells, were immunoprecipitated with antibody to FANCD2 (E35), FANCE, or a control nonimmunized rabbit serum (Pre-imm), and the immune complexes were immunoblotted with anti-BRCA2 (Ab-1) or anti-FANCD2 (FI-17) antibodies. Heavy chain IgG was used as a loading control. (E) Schematic model of the ATM-BRCA2-FANCD2-mediated DNA damage response. IR activates ATM, resulting in the phosphorylation of BRCA2, FANCD2, and several other protein substrates. Activated BRCA2 is then recruited to chromatin by FANCD2, which is activated by monoubiquitination. The interaction of BRCA2 and FANCD2 requires both the C terminus of BRCA2 and FANCD2 monoubiquitination. Following its loading onto chromatin, BRCA2 then functions downstream of monubiquitinated FANCD2.
Similar articles
- FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex.
Raghunandan M, Chaudhury I, Kelich SL, Hanenberg H, Sobeck A. Raghunandan M, et al. Cell Cycle. 2015;14(3):342-53. doi: 10.4161/15384101.2014.987614. Cell Cycle. 2015. PMID: 25659033 Free PMC article. - FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3.
Wilson JB, Yamamoto K, Marriott AS, Hussain S, Sung P, Hoatlin ME, Mathew CG, Takata M, Thompson LH, Kupfer GM, Jones NJ. Wilson JB, et al. Oncogene. 2008 Jun 12;27(26):3641-52. doi: 10.1038/sj.onc.1211034. Epub 2008 Jan 21. Oncogene. 2008. PMID: 18212739 - Direct interaction of the Fanconi anaemia protein FANCG with BRCA2/FANCD1.
Hussain S, Witt E, Huber PA, Medhurst AL, Ashworth A, Mathew CG. Hussain S, et al. Hum Mol Genet. 2003 Oct 1;12(19):2503-10. doi: 10.1093/hmg/ddg266. Epub 2003 Aug 5. Hum Mol Genet. 2003. PMID: 12915460 - The interplay of Fanconi anemia proteins in the DNA damage response.
Wang X, D'Andrea AD. Wang X, et al. DNA Repair (Amst). 2004 Aug-Sep;3(8-9):1063-9. doi: 10.1016/j.dnarep.2004.04.005. DNA Repair (Amst). 2004. PMID: 15279794 Review. - Regulation of the Fanconi anemia pathway by monoubiquitination.
Gregory RC, Taniguchi T, D'Andrea AD. Gregory RC, et al. Semin Cancer Biol. 2003 Feb;13(1):77-82. doi: 10.1016/s1044-579x(02)00102-5. Semin Cancer Biol. 2003. PMID: 12507559 Review.
Cited by
- The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI.
Williams SA, Longerich S, Sung P, Vaziri C, Kupfer GM. Williams SA, et al. Blood. 2011 May 12;117(19):5078-87. doi: 10.1182/blood-2010-10-311761. Epub 2011 Feb 25. Blood. 2011. PMID: 21355096 Free PMC article. - The carboxyl terminus of FANCE recruits FANCD2 to the Fanconi Anemia (FA) E3 ligase complex to promote the FA DNA repair pathway.
Polito D, Cukras S, Wang X, Spence P, Moreau L, D'Andrea AD, Kee Y. Polito D, et al. J Biol Chem. 2014 Mar 7;289(10):7003-7010. doi: 10.1074/jbc.M113.533976. Epub 2014 Jan 22. J Biol Chem. 2014. PMID: 24451376 Free PMC article. - The ups and downs of DNA repair biomarkers for PARP inhibitor therapies.
Wang X, Weaver DT. Wang X, et al. Am J Cancer Res. 2011;1(3):301-327. Epub 2010 Jan 3. Am J Cancer Res. 2011. PMID: 21968427 Free PMC article. - The NAE inhibitor pevonedistat interacts with the HDAC inhibitor belinostat to target AML cells by disrupting the DDR.
Zhou L, Chen S, Zhang Y, Kmieciak M, Leng Y, Li L, Lin H, Rizzo KA, Dumur CI, Ferreira-Gonzalez A, Rahmani M, Povirk L, Chalasani S, Berger AJ, Dai Y, Grant S. Zhou L, et al. Blood. 2016 May 5;127(18):2219-30. doi: 10.1182/blood-2015-06-653717. Epub 2016 Feb 5. Blood. 2016. PMID: 26851293 Free PMC article. - Guardians of the Genome: BRCA2 and Its Partners.
Le HP, Heyer WD, Liu J. Le HP, et al. Genes (Basel). 2021 Aug 10;12(8):1229. doi: 10.3390/genes12081229. Genes (Basel). 2021. PMID: 34440403 Free PMC article. Review.
References
- Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. - PubMed
- Andegeko, Y., L. Moyal, L. Mittelman, I. Tsarfaty, Y. Shiloh, and G. Rotman. 2001. Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem. 276:38224-38230. - PubMed
- Bruun, D., A. Folias, Y. M. Akkari, Y. Cox, S. Olson, and R. Moses. 2003. siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair 2:1007-1013. - PubMed
- Cortez, D., Y. Wang, J. Qin, and S. J. Elledge. 1999. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286:1162-1166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL052725/HL/NHLBI NIH HHS/United States
- P01HL54785/HL/NHLBI NIH HHS/United States
- P50 HL054785/HL/NHLBI NIH HHS/United States
- R01HL52725/HL/NHLBI NIH HHS/United States
- R01 DK043889/DK/NIDDK NIH HHS/United States
- R01DK43889/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous