High-resolution characterization of the pancreatic adenocarcinoma genome - PubMed (original) (raw)
. 2004 Jun 15;101(24):9067-72.
doi: 10.1073/pnas.0402932101.
Cameron Brennan, Gerald Bailey, Raktim Sinha, Bin Feng, Christopher Leo, Yunyu Zhang, Jean Zhang, Joseph D Gans, Nabeel Bardeesy, Craig Cauwels, Carlos Cordon-Cardo, Mark S Redston, Ronald A DePinho, Lynda Chin
Affiliations
- PMID: 15199222
- PMCID: PMC428474
- DOI: 10.1073/pnas.0402932101
High-resolution characterization of the pancreatic adenocarcinoma genome
Andrew J Aguirre et al. Proc Natl Acad Sci U S A. 2004.
Abstract
The pancreatic adenocarcinoma genome harbors multiple amplifications and deletions, pointing to the existence of numerous oncogenes and tumor suppressor genes driving the genesis and progression of this lethal cancer. Here, array comparative genomic hybridization on a cDNA microarray platform and informatics tools have been used to define the copy number alterations in a panel of 24 pancreatic adenocarcinoma cell lines and 13 primary tumor specimens. This high-resolution genomic analysis has identified all known regional gains and losses as well as many previously uncharacterized highly recurrent copy number alterations. A systematic prioritization scheme has selected 64 focal minimal common regions (MCRs) of recurrent copy number change. These MCRs possess a median size of 2.7 megabases (Mb), with 21 (33%) MCRs spanning 1 Mb or less (median of 0.33 Mb) and possessing an average of 15 annotated genes. Furthermore, complementary expression profile analysis of a significant fraction of the genes residing within these 64 prioritized MCRs has enabled the identification of a subset of candidates with statistically significant association between gene dosage and mRNA expression. Thus, the integration of DNA and RNA profiles provides a highly productive entry point for the discovery of genes involved in the pathogenesis of pancreatic adenocarcinoma.
Figures
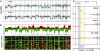
Fig. 1.
Genomic profiles from pancreatic adenocarcinoma samples. Array-CGH profiles with x axis coordinates representing cDNA probes ordered by genomic map positions. Segmented data are displayed in red, median filtered (three nearest neighbors) in blue, and raw data in black. (A) Whole-genome profiles of primary tumor specimen PA.T.7692 (Upper) and cell line Panc 10.05 (Lower). Note presence of focal high-level amplifications and deletions as well as large regional gains and losses in both samples. (B) Recurrence of chromosomal alterations. (Upper) Integer-value recurrence of CNAs in segmented data (y axis) plotted for each cDNA probe evenly aligned along the x axis in genome order. Dark red or green bars denote gain or loss of chromosome material. Bright red or green bars represent probes within regions of amplification or deletion. (Lower)
treeview
(Eisen laboratory, University of California, Berkeley) showing discrete CNAs within all samples. Red represents chromosomal gain, and green denotes chromosomal loss. (C) CGH profiles of 12p12.3-q13.3 locus (locus no. 15, Table 1) in three samples illustrating the definition of the physical extent, peak profile, and MCRs for that locus. Note that the left MCR is defined by the overlap between samples on top and bottom, whereas the right MCR is defined by the overlap between the two samples on top. Because data points are plotted on the x axis by genomic map positions, gaps in the profiles encompass regions of copy number transition for which there is no data point.
Fig. 2.
QPCR verifies complexity within CNAs. (A) Chromosome 7 CGH profiles (Left) showing amplification of a discrete region of 7q22 in both the AsPC-1 cell line and PA.T.14172 (locus no. 9, Table 1), with MCR defined by both samples (outlined by dashed lines). A-D indicate the relative positions of QPCR assays (Right), which confirm the gene copy alterations in AsPC-1 (dark gray bars) and PA.T.14172 (light gray bars). (B) Chromosome 9 array-CGH profile (Left) for a complex CNA in the HUP-T3 cell line. Homozygous deletion of the known target p16Ink4a is confirmed by QPCR (Right), which also verifies existence of two discrete focal amplicons and a narrow region of one-copy loss revealed by array-CGH. Note that CNAs covered by only one or two probes are not identified by the segmentation algorithm.
Fig. 3.
Combined array-CGH and expression analysis facilitates identification of candidate genes. (A) Analysis of 17q23.2-25.3 locus (locus no. 21, Table 1) in cell line Hup T3. (Upper) Array-CGH profile of HUP-T3. (Lower) Expression profile of genes on Affymetrix U133A array within the specified locus for the HUP-T3 cell line. Note that the subset of genes exhibiting prominent gene dosage correlated expression fall within the peak of the locus (arrows). (B) Analysis of 9p24.3-21.2 locus (locus no. 41, Table 1) in the cell line BxPC-3. (Upper) Array-CGH profile of the 9p region. (Lower) Affymetrix expression profile of genes mapping to the same region. Note the dramatically reduced expression of the p16INK4A gene (arrows) within the MCR. (C) Correlation of p16INK4A expression and copy number in 24 cell lines analyzed. Note the bimodal distribution of both expression values and copy number values for this gene across all samples (green lines). The red box defines those samples (BxPC-3, MiaPaCa, Capan 1, Hup-T3, and Dan-G) in which p16INK4A is homozygously deleted and not expressed. The blue box encloses samples (Panc-1, Panc 03.27, SW1990, Panc 08.13, Hup-T4, and Panc 02.13) in which p16INK4A is present but with absent or reduced expression.
Similar articles
- Genome-wide analysis of pancreatic cancer using microarray-based techniques.
Harada T, Chelala C, Crnogorac-Jurcevic T, Lemoine NR. Harada T, et al. Pancreatology. 2009;9(1-2):13-24. doi: 10.1159/000178871. Epub 2008 Dec 12. Pancreatology. 2009. PMID: 19077451 Review. - Pancreatic adenocarcinoma -- genetic portrait from chromosomes to microarrays.
Karhu R, Mahlamäki E, Kallioniemi A. Karhu R, et al. Genes Chromosomes Cancer. 2006 Aug;45(8):721-30. doi: 10.1002/gcc.20337. Genes Chromosomes Cancer. 2006. PMID: 16688744 Review. - Genomic DNA-chip hybridization reveals a higher incidence of genomic amplifications in pancreatic cancer than conventional comparative genomic hybridization and leads to the identification of novel candidate genes.
Holzmann K, Kohlhammer H, Schwaenen C, Wessendorf S, Kestler HA, Schwoerer A, Rau B, Radlwimmer B, Döhner H, Lichter P, Gress T, Bentz M. Holzmann K, et al. Cancer Res. 2004 Jul 1;64(13):4428-33. doi: 10.1158/0008-5472.CAN-04-0431. Cancer Res. 2004. PMID: 15231651 - High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization.
Jönsson G, Staaf J, Olsson E, Heidenblad M, Vallon-Christersson J, Osoegawa K, de Jong P, Oredsson S, Ringnér M, Höglund M, Borg A. Jönsson G, et al. Genes Chromosomes Cancer. 2007 Jun;46(6):543-58. doi: 10.1002/gcc.20438. Genes Chromosomes Cancer. 2007. PMID: 17334996 - Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex genetic alterations in cervical cancer.
Kloth JN, Oosting J, van Wezel T, Szuhai K, Knijnenburg J, Gorter A, Kenter GG, Fleuren GJ, Jordanova ES. Kloth JN, et al. BMC Genomics. 2007 Feb 20;8:53. doi: 10.1186/1471-2164-8-53. BMC Genomics. 2007. PMID: 17311676 Free PMC article.
Cited by
- Integrated high-resolution array CGH and SKY analysis of homozygous deletions and other genomic alterations present in malignant mesothelioma cell lines.
Klorin G, Rozenblum E, Glebov O, Walker RL, Park Y, Meltzer PS, Kirsch IR, Kaye FJ, Roschke AV. Klorin G, et al. Cancer Genet. 2013 May;206(5):191-205. doi: 10.1016/j.cancergen.2013.04.006. Epub 2013 Jul 5. Cancer Genet. 2013. PMID: 23830731 Free PMC article. - PTEN Alterations as a Potential Mechanism for Tumor Cell Escape from PD-1/PD-L1 Inhibition.
Cretella D, Digiacomo G, Giovannetti E, Cavazzoni A. Cretella D, et al. Cancers (Basel). 2019 Sep 6;11(9):1318. doi: 10.3390/cancers11091318. Cancers (Basel). 2019. PMID: 31500143 Free PMC article. Review. - Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome.
Loukopoulos P, Shibata T, Katoh H, Kokubu A, Sakamoto M, Yamazaki K, Kosuge T, Kanai Y, Hosoda F, Imoto I, Ohki M, Inazawa J, Hirohashi S. Loukopoulos P, et al. Cancer Sci. 2007 Mar;98(3):392-400. doi: 10.1111/j.1349-7006.2007.00395.x. Epub 2007 Jan 12. Cancer Sci. 2007. PMID: 17233815 Free PMC article. - Array-CGH reveals recurrent genomic changes in Merkel cell carcinoma including amplification of L-Myc.
Paulson KG, Lemos BD, Feng B, Jaimes N, Peñas PF, Bi X, Maher E, Cohen L, Leonard JH, Granter SR, Chin L, Nghiem P. Paulson KG, et al. J Invest Dermatol. 2009 Jun;129(6):1547-55. doi: 10.1038/jid.2008.365. Epub 2008 Nov 20. J Invest Dermatol. 2009. PMID: 19020549 Free PMC article. - A novel method, digital genome scanning detects KRAS gene amplification in gastric cancers: involvement of overexpressed wild-type KRAS in downstream signaling and cancer cell growth.
Mita H, Toyota M, Aoki F, Akashi H, Maruyama R, Sasaki Y, Suzuki H, Idogawa M, Kashima L, Yanagihara K, Fujita M, Hosokawa M, Kusano M, Sabau SV, Tatsumi H, Imai K, Shinomura Y, Tokino T. Mita H, et al. BMC Cancer. 2009 Jun 23;9:198. doi: 10.1186/1471-2407-9-198. BMC Cancer. 2009. PMID: 19545448 Free PMC article.
References
- Bardeesy, N. & DePinho, R. A. (2002) Nat. Rev. Cancer 2, 897-909. - PubMed
- Johansson, B., Bardi, G., Heim, S., Mandahl, N., Mertens, F., Bak-Jensen, E., Andren-Sandberg, A. & Mitelman, F. (1992) Cancer 69, 1674-1681. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA99041/CA/NCI NIH HHS/United States
- R01 CA099041/CA/NCI NIH HHS/United States
- T32 CA09382/CA/NCI NIH HHS/United States
- R01 CA086379/CA/NCI NIH HHS/United States
- T32 CA009382/CA/NCI NIH HHS/United States
- R01CA86379/CA/NCI NIH HHS/United States
- R01 CA084628/CA/NCI NIH HHS/United States
- R01CA84628/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous