Displacements of prohead protease genes in the late operons of double-stranded-DNA bacteriophages - PubMed (original) (raw)
Displacements of prohead protease genes in the late operons of double-stranded-DNA bacteriophages
Jing Liu et al. J Bacteriol. 2004 Jul.
Abstract
Most of the known prohead maturation proteases in double-stranded-DNA bacteriophages are shown, by computational methods, to fall into two evolutionarily independent clans of serine proteases, herpesvirus assemblin-like and ClpP-like. Phylogenetic analysis suggests that these two types of phage prohead protease genes displaced each other multiple times while preserving their exact location within the late operons of the phage genomes.
Figures
FIG. 1.
Structures of HCMV protease (PDB code 1CMV) and ClpP protease from E. coli (PDB code 1TYF). The α-helices are blue, and the β-strands are orange. The residues in the catalytic triad are shown with balls and sticks. They are His63-Ser132-His157 in HCMV protease and Ser97-His122-Asp171 in E. coli ClpP protease. The structures were drawn by using the program MOLSCRIPT (19).
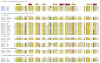
FIG. 2.
Multiple-sequence alignment of protease clan SH members. The left column is the gi number for each sequence followed by the name of the virus or phage. The identifiers of herpesvirus proteases with known three-dimensional structures are shown in bold blue font. Distances, in amino acid residues, from the ends of each sequence and between the blocks with highest sequence similarities are shown in parentheses. Consensus positions of the structural elements are shown above the alignment. Yellow shading indicates the conservation of hydrophobic residues, gray shading indicates the conservation of residues with small side chains (A, G, and S), and a white font on a black background indicates the conservation of negatively charged residues (D and E). The catalytic Ser, His, and Asp/Glu residues in the catalytic triad are in white font on a red background, except that the His in the third position of the triad in all herpesvirus proteases is on a blue background. (A) Representatives from family S21 with known three-dimensional structures. HSV-2, herpes simplex virus 2; VZV, varicella-zoster virus; EBV, Epstein-Barr virus; KSHV, Kaposi's sarcoma-associated herpesvirus. (B) Representatives from family U35. (C) Representatives from family U9.
FIG. 3.
Multiple-sequence alignment of protease clan SK members. Designations are as described in the legend to Fig. 2. Species abbreviations: Ec, E. coli; Bs, Bacillus subtilis; Lp, Lactobacillus plantarum; At, Arabidopsis thaliana; Sco, Streptomyces coelicolor A3(2); Sy, Synechocystis sp. strain PCC 6803; Bh, Bacillus halodurans; Pf, Pyrococcus furiosus DSM 3638; Mj, Methanococcus jannaschii; Rc, Rickettsia conorii.
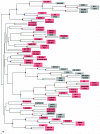
FIG. 4.
Phylogenetic tree constructed by using the sequence alignment of phage portal proteins. Sequences of the portal proteins from phages and prophages were aligned by using the T-Coffee (23) and CLUSTAL_X (34) programs, followed by manual validation and the removal of poorly aligned regions. The phylogenetic tree was constructed by using the neighbor-joining method as implemented in the NEIGHBOR program, and subsets of the data were used to rebuild the tree multiple times. The root was set to midpoint by the RETREE program of the PHYLIP package (10), and the consensus tree was subsequently reviewed by TreeView (24). Bootstrap values were estimated by resampling the set of the alignment 100 times (see the supplemental material). The level of bootstrap support is marked by small circles in the following colors: red (90 to 100%), yellow (80 to 90%), green (70 to 80%), and blue (50 to 70%). The nodes with <40% support are unlabeled. Phages that encode prohead protease belonging to clan SH are shaded in pink, and those that encode prohead protease belonging to clan SK are shaded in gray.
Similar articles
- Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease.
Cheng H, Shen N, Pei J, Grishin NV. Cheng H, et al. Protein Sci. 2004 Aug;13(8):2260-9. doi: 10.1110/ps.04726004. Protein Sci. 2004. PMID: 15273316 Free PMC article. - A snapshot of viral evolution from genome analysis of the tectiviridae family.
Saren AM, Ravantti JJ, Benson SD, Burnett RM, Paulin L, Bamford DH, Bamford JK. Saren AM, et al. J Mol Biol. 2005 Jul 15;350(3):427-40. doi: 10.1016/j.jmb.2005.04.059. J Mol Biol. 2005. PMID: 15946683 - The 1.2-megabase genome sequence of Mimivirus.
Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. Raoult D, et al. Science. 2004 Nov 19;306(5700):1344-50. doi: 10.1126/science.1101485. Epub 2004 Oct 14. Science. 2004. PMID: 15486256 - Protein repertoire of double-stranded DNA bacteriophages.
Liu J, Glazko G, Mushegian A. Liu J, et al. Virus Res. 2006 Apr;117(1):68-80. doi: 10.1016/j.virusres.2006.01.015. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16490276 Review. - Mimivirus and the emerging concept of "giant" virus.
Claverie JM, Ogata H, Audic S, Abergel C, Suhre K, Fournier PE. Claverie JM, et al. Virus Res. 2006 Apr;117(1):133-44. doi: 10.1016/j.virusres.2006.01.008. Epub 2006 Feb 15. Virus Res. 2006. PMID: 16469402 Review.
Cited by
- Characterization of the temperate phage vB_RleM_PPF1 and its site-specific integration into the Rhizobium leguminosarum F1 genome.
Halmillawewa AP, Restrepo-Córdoba M, Perry BJ, Yost CK, Hynes MF. Halmillawewa AP, et al. Mol Genet Genomics. 2016 Feb;291(1):349-62. doi: 10.1007/s00438-015-1113-8. Epub 2015 Sep 16. Mol Genet Genomics. 2016. PMID: 26377943 - Common Evolutionary Origin of Procapsid Proteases, Phage Tail Tubes, and Tubes of Bacterial Type VI Secretion Systems.
Fokine A, Rossmann MG. Fokine A, et al. Structure. 2016 Nov 1;24(11):1928-1935. doi: 10.1016/j.str.2016.08.013. Epub 2016 Sep 22. Structure. 2016. PMID: 27667692 Free PMC article. - Polyvalent Proteins, a Pervasive Theme in the Intergenomic Biological Conflicts of Bacteriophages and Conjugative Elements.
Iyer LM, Burroughs AM, Anand S, de Souza RF, Aravind L. Iyer LM, et al. J Bacteriol. 2017 Jul 11;199(15):e00245-17. doi: 10.1128/JB.00245-17. Print 2017 Aug 1. J Bacteriol. 2017. PMID: 28559295 Free PMC article. - Sequencing Bacillus anthracis typing phages gamma and cherry reveals a common ancestry.
Fouts DE, Rasko DA, Cer RZ, Jiang L, Fedorova NB, Shvartsbeyn A, Vamathevan JJ, Tallon L, Althoff R, Arbogast TS, Fadrosh DW, Read TD, Gill SR. Fouts DE, et al. J Bacteriol. 2006 May;188(9):3402-8. doi: 10.1128/JB.188.9.3402-3408.2006. J Bacteriol. 2006. PMID: 16621835 Free PMC article. - Complete genome of the broad-host-range Erwinia amylovora phage phiEa21-4 and its relationship to Salmonella phage felix O1.
Lehman SM, Kropinski AM, Castle AJ, Svircev AM. Lehman SM, et al. Appl Environ Microbiol. 2009 Apr;75(7):2139-47. doi: 10.1128/AEM.02352-08. Epub 2009 Jan 30. Appl Environ Microbiol. 2009. PMID: 19181832 Free PMC article.
References
- Black, L. W., and M. K. Showe. 1983. Morphogenesis of the T4 head, p. 219-245. In C. K. Mathews, E. M. Kutter, G. Mosig, and P. B. Berget (ed.), Bacteriophage T4. American Society for Microbiology, Washington, D.C.
- Buisson, M., J. F. Hernandez, D. Lascoux, G. Schoehn, E. Forest, G. Arlaud, J. M. Seigneurin, R. W. Ruigrok, and W. P. Burmeister. 2002. The crystal structure of the Epstein-Barr virus protease shows rearrangement of the processed C terminus. J. Mol. Biol. 324:89-103. - PubMed
- Casjens, S., G. Hatfull, and R. Hendrix. 1992. Evolution of the dsDNA tailed-bacteriophage genomes. Semin. Virol. 3:383-397.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources