Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1 - PubMed (original) (raw)
Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1
Andreas Marg et al. J Cell Biol. 2004.
Abstract
Interferon stimulation of cells leads to the tyrosine phosphorylation of latent Stat1 and subsequent transient accumulation in the nucleus that requires canonical transport factors. However, the mechanisms that control the predominantly cytoplasmic localization in unstimulated cells have not been resolved. We uncovered that constitutive energy- and transport factor-independent nucleocytoplasmic shuttling is a property of unphosphorylated Stat1, Stat3, and Stat5. The NH(2)- and COOH-terminal Stat domains are generally dispensable, whereas alkylation of a single cysteine residue blocked cytokine-independent nuclear translocation and thus implicated the linker domain into the cycling of Stat1. It is revealed that constitutive nucleocytoplasmic shuttling of Stat1 is mediated by direct interactions with the FG repeat regions of nucleoporin 153 and nucleoporin 214 of the nuclear pore. Concurrent active nuclear export by CRM1 created a nucleocytoplasmic Stat1 concentration gradient that is significantly reduced by the blocking of energy-requiring translocation mechanisms or the specific inactivation of CRM1. Thus, we propose that two independent translocation pathways cooperate to determine the steady-state distribution of Stat1.
Figures
Figure 1.
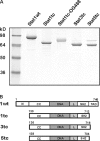
Recombinant Stat proteins. (A) SDS-PAGE (7%) analysis of the Stat protein preparations used in this work (1 μg of each). Molecular size markers are indicated to the left. OG488, Oregon green 488. (B) Schematic representation of the respective Stat proteins. The truncated mutants were expressed with a COOH-terminal Strep-tag of the sequence SAWSHPQFEK (single letter code). The Stat proteins were of human (Stat1), mouse (Stat3), or sheep (Stat5) origin. N, N domain; CC, coiled coil domain; DNA, DNA-binding domain; L, linker domain; SH2, SH2 domain; TAD, transactivation domain.
Figure 2.
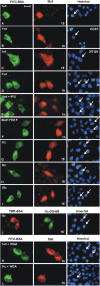
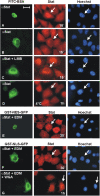
Unphosphorylated recombinant Stat proteins translocate into the nucleus of unstimulated cells. HeLa-S3 cells (except for B and C) were used. The injection site is indicated by the location of co-microinjected FITC-labeled or tetramethyl-rhodamine (TMR)–labeled BSA. Nuclei were stained with Hoechst dye. Arrows indicate injected cells. (A–C) Cytoplasmic microinjection of purified Stat1wt into HeLa cells (A) , COS7 cells (B), or 2fTGH cells (C). After 15 min the cells were fixed and Stat1 was detected by immunocytochemistry with a specific antibody. (D) Identical experimental setup as in A, but microinjected cells were fixed after 60 min (E). Identical to D, but cells were treated with IFNγ. (F–I) Cytoplasmic microinjection of full-length tyrosine mutant Y701F (F), or truncated Stat1tc (G), Stat3tc (H), or Stat5tc (I). Fixation was 15 min after injection, truncated Stats were detected with Strep-tag antibody. (J) Cytoplasmic microinjection of Oregon green 488–labeled Stat1tc. Cells were fixed after 15 min followed by direct fluorescence microscopy. (K and L) Cytoplasmic co-microinjection of WGA and Stat1wt (K) or Stat1tc (L). After 1 h the cells were fixed and Stat1 variant proteins were detected by immunocytochemistry as described. Bar, 20 μm.
Figure 3.
Unphosphorylated recombinant Stat proteins exit the nucleus of unstimulated cells. The injection sites of the HeLa cells are indicated by co-microinjected FITC-labeled BSA. (A and B) Recombinant Stat1wt was injected into the nucleus. After 1 h (A) or 3 h (B) immunocytochemistry with a Stat1-specific antibody was performed on fixed cells. (C and D) Microinjection of Stat1wt into cells pretreated for 1 h with LMB. After nuclear (closed arrow) or cytoplasmic (open arrow) microinjection the cells were further incubated with LMB for 1 h (C) or 4 h (D) before fixation and immunocytochemistry with a Stat1-specific antibody. (E and F) Stat1tc was injected into the nucleus and the cells were subsequently incubated for 1 h (E) or 4 h (F), before fixation and immunocytochemistry with Strep-tag antibody. (G) Nuclear microinjection of Alexa Fluor 594–labeled Stat1wt, followed by incubation for 2 h, fixation, and direct fluorescence microscopy. Bar, 20 μm.
Figure 4.
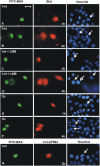
Unphosphorylated recombinant Stat proteins enter the nucleus of energy-depleted cells. HeLa cells were kept in growth medium or EDM for 2 h before microinjection. (A and B) Tyrosine-phosphorylated Stat1wt was injected into the cytoplasm of cells treated with growth medium (A) or EDM (B). (C–F) Unphosphorylated Stat1wt (C), Stat1tc (D), Stat3tc (E), or Stat5tc (F) were injected into the cytoplasm of cells incubated in EDM. Injection site and energy depletion are indicated by the location of co-microinjected GST-NLS-GFP. In A, FITC-BSA was used as the injection site marker. The cells were fixed 30 min after injection and Stats were detected by immunocytochemistry. (G) Nuclear co-microinjection of recombinant Stat1wt and GST-NES-GFP, followed by 3 h of incubation in EDM. Then, cells were processed for immunocytochemistry as before. Arrows indicate nuclei of microinjected cells. Bar, 20 μm.
Figure 5.
Nucleocytoplasmic translocation of endogenous Stat1 in normal and energy-depleted HeLa cells. Stat1 antibodies were microinjected followed by immunocytochemical detection of Stat1. (A) Microinjection of Stat1 antibodies into the cell nucleus with fixation after 15 min. The injection site is marked by co-microinjected FITC-BSA. (B) Identical with A, except that the antibodies were microinjected into the cytoplasm. (C) Identical with B except that the cells were incubated with 5 ng/ml LMB starting 1 h before microinjection. (D) Identical with B, except that the cells were kept at 4° C for 60 min both before and after cytoplasmic microinjection. (E) Stat1 antibodies were microinjected into the nucleus of cells after a 2-h preincubation in EDM. Injection site and energy depletion are indicated by co-microinjected GST-NES-GFP. The cells were fixed 30 min after microinjection. (F and G) Cytoplasmic microinjection of Stat1 antibody in energy-depleted cells. WGA was co-microinjected in G. Injection site and energy depletion are indicated by co-microinjected GST-NLS-GFP. Arrows indicate positions of microinjected cells. Bar, 20 μm.
Figure 6.
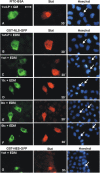
The nuclear import of unphosphorylated Stat proteins continues in cytosol-depleted digitonin-permeabilized cells. HeLa-S3 cells were treated with 40 μg/ml digitonin, washed, and incubated with 20 μl of IM (protein-free TB supplemented with 10 mg/ml BSA) at RT unless noted otherwise. The nuclei were stained with Hoechst dye. Unlabeled Stat proteins were detected immunocytochemically in fixed cells. (A) Permeabilized HeLa cells were incubated for 60 min with 1 μM unphosphorylated Stat1tc in IM. In addition, the import reaction was performed on ice (4°C), or after preincubation with 250 μg/ml WGA (+WGA). Where indicated, the following additions were made to IM. Rabbit reticulocyte lysate (75%) and an energy regeneration system (+CIM); 0.8 U/ml apyrase (+apyrase); 16 μM truncated p97 (residues 45–462) fused to GST (+p97tc); 20 μM Stat1wt (+1wt). (B) Import reaction with tyrosine-phosphorylated Stat1wt in CIM or IM. (C) Identical experimental setup as in A, but with Stat3tc replacing Stat1tc. (D–G) Time course of carrier-free nuclear import of Stat1wt (D), Stat1tc (E), Stat3tc (F), or Oregon green 488–labeled Stat1tc (G). The IM contained 1 μM of the respective Stat proteins in IM. In G Stat1tc was detected by direct fluorescence microscopy. Note the different time scale. Bar, 30 μm.
Figure 7.
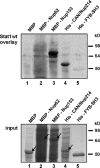
Stat1 interacts with FG repeat–containing Nups. (Top) Bacterial lysates containing the indicated fusion proteins (lanes 1–4) or purified His-tagged tandem SH3 domains from FYB (lane 5) were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with wild-type unphosphorylated Stat1. The association of Stat1 with the immobilized proteins was detected by immunoblotting using a Stat1-specific antibody. The bottom panel shows the respective Coomassie-blue stained SDS-gel before transfer to a nitrocellulose membrane (input). Arrows indicate the proteins listed above the respective lane.
Figure 8.
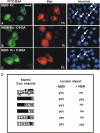
Alkylation of cysteine 543 in the linker domain precludes nuclear import of Stat1. (A–C) Purified Stat1tc or the indicated mutants were alkylated by NEM in vitro and injected in the cytosol of HeLa cells. After 1 h the cells were fixed and the intracellular distribution of the microinjected proteins was determined by immunocytochemistry. Arrows indicate nuclei of microinjected cells. Bar, 20 μm. (D) Summary of the microinjection data and mass spectrometric analyses of alkylated Stat1. Purified bacterially expressed Stat1tc was treated without or with NEM (Materials and methods) and subsequently injected into the cytoplasm of HeLa cells. The ability to enter the nucleus during a 1-h incubation period is stated. The cysteine residues in position 155, 174, 247, 255, 324, 440, 492, 543, 577 of human Stat1 are numbered 1 to 9 in the diagram to the left. Mutated cysteines (Cys to Ala) are blackened. Open circles denote absence of alkylation; closed circles denote alkylated cysteines as determined by mass spectrometry. For position 7 both alkylated and nonalkylated peptides were found. Peptides covering Cys-positions 3, 4, and 6 were not detected.
Figure 9.
The inhibition of active nuclear export prevents cytoplasmic accumulation of Stat1. (A) 3T3 cells were left untreated (DMEM) or treated with EDM for 2 h followed by growth medium for 5 h (EDM+DMEM). Alternatively, cells were treated with the CRM1 inhibitor leptomycin B (LMB) for 2 or 5 h as indicated. Shown is the distribution of the endogenous Stat1 as revealed by immunocytochemistry with a specific antibody and conventional microscopy. Bar, 20 μm. (B) Corresponding bar diagram with a quantitative analysis of the ratio of the mean cytoplasmic and nuclear immunofluorescence densities. The median slice (x/y image) of a confocal microscopical image was used to quantify the signals in 13 randomly chosen cells Values are expressed as mean ± SD. Statistically significant differences between cells incubating in control (DMEM) or experimental medium are indicated by the asterisks.
Figure 10.
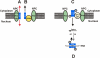
A model of Stat1 nucleocytoplasmic shuttling. (A) Unphosphorylated “latent” Stat1 constitutively shuttles between cytosol and nucleoplasm via direct interactions with the Nup153 and Nup214. The surface of the linker domain of Stat1 is likely to provide the contact surface. (B) In addition, NES-mediated transport of unphosphorylated Stat1 via CRM1 enhances the export rate and achieves cytoplasmic accumulation. (C) After cytokine-induced receptor activation, Stat1 is tyrosine phosphorylated and dimerizes, which precludes further carrier-free nucleocytoplasmic cycling. However, a dimer-specific NLS in the DNA-binding domain is exposed, and nuclear import occurs in complex with NPI-1 and p97. (D) Until its dephosphorylation, which is inhibited by DNA-binding, Stat1 is retained in the nucleus, thus allowing for the signal-induced nuclear accumulation. GAS, Stat1-binding site.
Similar articles
- IFN-type-I-mediated signaling is regulated by modulation of STAT2 nuclear export.
Frahm T, Hauser H, Köster M. Frahm T, et al. J Cell Sci. 2006 Mar 15;119(Pt 6):1092-104. doi: 10.1242/jcs.02822. Epub 2006 Feb 28. J Cell Sci. 2006. PMID: 16507591 - Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFbeta signaling complexes in the cytoplasm and nucleus.
Xu L, Kang Y, Cöl S, Massagué J. Xu L, et al. Mol Cell. 2002 Aug;10(2):271-82. doi: 10.1016/s1097-2765(02)00586-5. Mol Cell. 2002. PMID: 12191473 - The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NFkappaB activation in Drosophila.
Xylourgidis N, Roth P, Sabri N, Tsarouhas V, Samakovlis C. Xylourgidis N, et al. J Cell Sci. 2006 Nov 1;119(Pt 21):4409-19. doi: 10.1242/jcs.03201. Epub 2006 Oct 10. J Cell Sci. 2006. PMID: 17032737 - The ins and outs of STAT1 nuclear transport.
McBride KM, Reich NC. McBride KM, et al. Sci STKE. 2003 Aug 12;2003(195):RE13. doi: 10.1126/stke.2003.195.re13. Sci STKE. 2003. PMID: 12915721 Review. - Nucleoporins and nucleocytoplasmic transport in hematologic malignancies.
Takeda A, Yaseen NR. Takeda A, et al. Semin Cancer Biol. 2014 Aug;27:3-10. doi: 10.1016/j.semcancer.2014.02.009. Epub 2014 Mar 18. Semin Cancer Biol. 2014. PMID: 24657637 Review.
Cited by
- FRAP analysis of nucleocytoplasmic dynamics of the vitamin D receptor splice variant VDRB1: preferential targeting to nuclear speckles.
Sunn KL, Eisman JA, Gardiner EM, Jans DA. Sunn KL, et al. Biochem J. 2005 Jun 1;388(Pt 2):509-14. doi: 10.1042/BJ20042040. Biochem J. 2005. PMID: 15689185 Free PMC article. - High Mobility Group B Proteins, Their Partners, and Other Redox Sensors in Ovarian and Prostate Cancer.
Barreiro-Alonso A, Lamas-Maceiras M, Rodríguez-Belmonte E, Vizoso-Vázquez Á, Quindós M, Cerdán ME. Barreiro-Alonso A, et al. Oxid Med Cell Longev. 2016;2016:5845061. doi: 10.1155/2016/5845061. Epub 2015 Nov 23. Oxid Med Cell Longev. 2016. PMID: 26682011 Free PMC article. Review. - Stat1 nuclear translocation by nucleolin upon monocyte differentiation.
Jerke U, Tkachuk S, Kiyan J, Stepanova V, Kusch A, Hinz M, Dietz R, Haller H, Fuhrman B, Dumler I. Jerke U, et al. PLoS One. 2009 Dec 14;4(12):e8302. doi: 10.1371/journal.pone.0008302. PLoS One. 2009. PMID: 20011528 Free PMC article. - Model-based extension of high-throughput to high-content data.
Pfeifer AC, Kaschek D, Bachmann J, Klingmüller U, Timmer J. Pfeifer AC, et al. BMC Syst Biol. 2010 Aug 5;4:106. doi: 10.1186/1752-0509-4-106. BMC Syst Biol. 2010. PMID: 20687942 Free PMC article. - Structural and functional studies of STAT1 from Atlantic salmon (Salmo salar).
Skjesol A, Hansen T, Shi CY, Thim HL, Jørgensen JB. Skjesol A, et al. BMC Immunol. 2010 Mar 30;11:17. doi: 10.1186/1471-2172-11-17. BMC Immunol. 2010. PMID: 20353564 Free PMC article.
References
- Darnell, J.E., Jr. 1997. Stats and gene regulation. Science. 277:1630–1635. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous