Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity - PubMed (original) (raw)
Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity
Ursula J Buchholz et al. Proc Natl Acad Sci U S A. 2004.
Abstract
We investigated the contributions of the structural proteins of severe acute respiratory syndrome (SARS) coronavirus (CoV) to protective immunity by expressing them individually and in combinations from a recombinant parainfluenza virus (PIV) type 3 vector called BHPIV3. This vector provided direct immunization of the respiratory tract, the major site of SARS transmission, replication, and disease. The BHPIV3/SARS recombinants were evaluated for immunogenicity and protective efficacy in hamsters, which support a high level of pulmonary SARS-CoV replication. A single intranasal administration of BHPIV3 expressing the SARS-CoV spike protein (S) induced a high titer of SARS-CoV-neutralizing serum antibodies, only 2-fold less than that induced by SARS-CoV infection. The expression of S with the two other putative virion envelope proteins, the matrix M and small envelope E proteins, did not augment the neutralizing antibody response. In absence of S, expression of M and E or the nucleocapsid protein N did not induce a detectable serum SARS-CoV-neutralizing antibody response. Immunization with BHPIV3 expressing S provided complete protection against SARS-CoV challenge in the lower respiratory tract and partial protection in the upper respiratory tract. This was augmented slightly by coexpression with M and E. Expression of M, E, or N in the absence of S did not confer detectable protection. These results identify S among the structural proteins as the only significant SARS-CoV neutralization antigen and protective antigen and show that a single mucosal immunization is highly protective in an experimental animal that supports efficient replication of SARS-CoV.
Figures
Fig. 1.
Maps of the genomes of BHPIV3 vectors expressing the SARS-CoV S, M, E, and N ORFs or a heterologous control sequence equivalent in length to the S ORF. The BPIV3 backbone genes are shown as open rectangles, the HPIV3 surface protein genes are checked rectangles, and the SARS-CoV ORFs and control sequence are shaded rectangles. PIV3 gene-start and gene-end transcription signals are shown as black triangles and bars, respectively, flanking each rectangle. The figure is not drawn to scale. le, leader; tr, trailer.
Fig. 2.
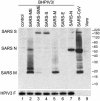
Transcription of the SARS-CoV inserts into monocistronic mRNAs. LLC-MK2 cells were mock-infected (lane 7) or infected with the indicated BHPIV3 vector (lanes 1-6) at an input multiplicity of infection of 3 TCID50 per cell. Total intracellular RNA was isolated 72 h after infection, electrophoresed on a 1% agarose gel in the presence of 0.44 M formaldehyde, transferred to nylon membrane, and hybridized with α-32P-labeled double-stranded DNA probes specific for SARS S, M, E, and N, or BPIV3 N, as indicated on the left. The images were prepared with a PhosphorImager.
Fig. 3.
Expression of SARS-CoV proteins by BHPIV3 vectors. LLC-MK2 cells were infected with the individual BHPIV3 vectors (lanes 1-7) at an input multiplicity of infection of 5 TCID50 per cell. In addition, Vero cells were mock-infected (lane 9) or infected with 5 TCID50 per cell of SARS-CoV (lane 8). Cell lysates were prepared 42 h after infection, in the case of SARS-CoV, or 72 h after infection, in the case of the BHPIV3 vectors, denatured under reducing conditions, subjected to SDS/PAGE in 4-12% gradient gels, and transferred to nitrocellulose. SARS-CoV proteins were detected by using convalescent serum from an African green monkey that had been infected with SARS-CoV. The BHPIV3 F protein was detected in a parallel blot by using HPIV3-specific rabbit hyperimmune serum (Lower). Detection of bound antibodies was done with horseradish peroxidase-conjugated goat anti-human (in the case of the anti-SARS-CoV serum) or anti-rabbit (in the case of the anti-HPIV3 serum) antibody, respectively, and visualized by chemiluminescence.
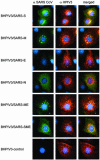
Fig. 4.
Indirect immunofluorescence of LLC-MK2 cells infected with BHPIV3 vectors. Cells on cover slips were infected at an input multiplicity of infection of 0.05 TCID50 with the BHPIV3 vectors listed to the left, incubated for 24 h, fixed with 4% paraformaldehyde, and permeabilized with 1% Triton X-100. SARS-CoV proteins were visualized (Left) by incubation with convalescent serum from a SARS-CoV-infected African green monkey followed by an Alexa488-conjugated goat anti-human antibody (Molecular Probes). BHPIV3 proteins were visualized (Center) by incubation with convalescence serum from HPIV3-infected hamsters, followed by Alexa594-conjugated goat anti-hamster antibody (Molecular Probes). Nuclear chromatin staining (blue) was performed with 4′,6-diamidino-2-phenylindole (Sigma).
Similar articles
- Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS.
Bukreyev A, Lamirande EW, Buchholz UJ, Vogel LN, Elkins WR, St Claire M, Murphy BR, Subbarao K, Collins PL. Bukreyev A, et al. Lancet. 2004 Jun 26;363(9427):2122-7. doi: 10.1016/S0140-6736(04)16501-X. Lancet. 2004. PMID: 15220033 Free PMC article. - Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus.
See RH, Zakhartchouk AN, Petric M, Lawrence DJ, Mok CPY, Hogan RJ, Rowe T, Zitzow LA, Karunakaran KP, Hitt MM, Graham FL, Prevec L, Mahony JB, Sharon C, Auperin TC, Rini JM, Tingle AJ, Scheifele DW, Skowronski DM, Patrick DM, Voss TG, Babiuk LA, Gauldie J, Roper RL, Brunham RC, Finlay BB. See RH, et al. J Gen Virol. 2006 Mar;87(Pt 3):641-650. doi: 10.1099/vir.0.81579-0. J Gen Virol. 2006. PMID: 16476986 - Chimeric severe acute respiratory syndrome coronavirus (SARS-CoV) S glycoprotein and influenza matrix 1 efficiently form virus-like particles (VLPs) that protect mice against challenge with SARS-CoV.
Liu YV, Massare MJ, Barnard DL, Kort T, Nathan M, Wang L, Smith G. Liu YV, et al. Vaccine. 2011 Sep 2;29(38):6606-13. doi: 10.1016/j.vaccine.2011.06.111. Epub 2011 Jul 14. Vaccine. 2011. PMID: 21762752 Free PMC article. - Vaccine design for severe acute respiratory syndrome coronavirus.
He Y, Jiang S. He Y, et al. Viral Immunol. 2005;18(2):327-32. doi: 10.1089/vim.2005.18.327. Viral Immunol. 2005. PMID: 16035944 Review. - Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus.
Cavanagh D. Cavanagh D. Avian Pathol. 2003 Dec;32(6):567-82. doi: 10.1080/03079450310001621198. Avian Pathol. 2003. PMID: 14676007 Free PMC article. Review.
Cited by
- Microstructure, pathophysiology, and potential therapeutics of COVID-19: A comprehensive review.
Singh SP, Pritam M, Pandey B, Yadav TP. Singh SP, et al. J Med Virol. 2021 Jan;93(1):275-299. doi: 10.1002/jmv.26254. Epub 2020 Jul 15. J Med Virol. 2021. PMID: 32617987 Free PMC article. Review. - An overview of some potential immunotherapeutic options against COVID-19.
Bayat M, Asemani Y, Mohammadi MR, Sanaei M, Namvarpour M, Eftekhari R. Bayat M, et al. Int Immunopharmacol. 2021 Jun;95:107516. doi: 10.1016/j.intimp.2021.107516. Epub 2021 Feb 26. Int Immunopharmacol. 2021. PMID: 33765610 Free PMC article. Review. - The Virological, Immunological, and Imaging Approaches for COVID-19 Diagnosis and Research.
Tan AS, Nerurkar SN, Tan WCC, Goh D, Lai CPT, Poh Sheng Yeong J. Tan AS, et al. SLAS Technol. 2020 Dec;25(6):522-544. doi: 10.1177/2472630320950248. Epub 2020 Aug 18. SLAS Technol. 2020. PMID: 32808850 Free PMC article. Review. - Vaccine design based on 16 epitopes of SARS-CoV-2 spike protein.
He J, Huang F, Zhang J, Chen Q, Zheng Z, Zhou Q, Chen D, Li J, Chen J. He J, et al. J Med Virol. 2021 Apr;93(4):2115-2131. doi: 10.1002/jmv.26596. Epub 2020 Nov 1. J Med Virol. 2021. PMID: 33091154 Free PMC article. - A combined nucleocapsid vaccine induces vigorous SARS-CD8+ T-cell immune responses.
Azizi A, Aucoin S, Tadesse H, Frost R, Ghorbani M, Soare C, Naas T, Diaz-Mitoma F. Azizi A, et al. Genet Vaccines Ther. 2005 Aug 22;3:7. doi: 10.1186/1479-0556-3-7. Genet Vaccines Ther. 2005. PMID: 16115319 Free PMC article.
References
- World Health Organization (2003) Wkly. Epidemiol. Rec. 78, 81-83. - PubMed
- Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Penaranda, S., Bankamp, B., Maher, K., Chen, M. H., et al. (2003) Science 300, 1394-1399. - PubMed
- Marra, M. A., Jones, S. J., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S., Khattra, J., Asano, J. K., Barber, S. A., Chan, S. Y., et al. (2003) Science 300, 1399-1404. - PubMed
- Lai, M. M. & Holmes, K. V. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), Vol. 2, pp. 1163-1203.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous