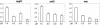Use of a continuous-flow anaerobic culture to characterize enteric virulence gene expression - PubMed (original) (raw)
Use of a continuous-flow anaerobic culture to characterize enteric virulence gene expression
Fernando Ruiz-Perez et al. Infect Immun. 2004 Jul.
Abstract
We developed an in vitro culture method to characterize the expression of bacterial genes under conditions mimicking the colonic environment. Our culture system (the intestinal simulator) comprised a continuous-flow anaerobic culture which was inoculated with fecal samples from healthy volunteers. As a test organism, we employed enteroaggregative Escherichia coli (EAEC), an emerging diarrheal pathogen that is thought to cause infection in both the small and large intestines. After the simulator culture achieved equilibrium conditions, we inoculated the system with prototype EAEC strain 042 and assessed the expression of three EAEC virulence-related genes. We focused particularly on expression of aggR, which encodes a global transcriptional regulator of EAEC virulence factors, and two AggR-regulated genes. By using real-time quantitative reverse transcription-PCR, we showed that aggR expression in the simulator is increased 3- to 10-fold when 042 is grown under low-pH (5.5 to 6.0) conditions, compared with results with neutral pH (7.0). Interestingly, however, this effect was seen only when the strain was grown in the presence of commensal bacteria. We also found that expression of aggR is 10- to 20-fold higher at low NaCl concentrations, and this effect was also observed only in the presence of commensal bacteria. Using coculture and conditioned-media experiments, we identified specific strains of Enterococcus and Clostridium that upregulated aggR expression; in contrast, strains of Lactobacillus and Veillonella downregulated aggR expression. Our data provide new insights into regulation of virulence genes in EAEC and suggest the utility of intestinal simulation cultures in characterizing enteric gene regulation.
Figures
FIG. 1.
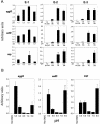
Main bacterial populations present in precultures of stool samples and after 7 days' growth in the simulator. The simulator culture was inoculated with 60 ml of precultured stool from a single subject and was incubated under continuous flow for 7 days. The dominant culturable bacterial populations were determined by biochemical analysis. The results for three representative experiments (S-1, S-2, and S-3) are shown.
FIG. 2.
Dominant culturable bacterial populations present in the simulator at different pH setpoints. EAEC 042 was inoculated in the intestinal simulator, and the system was set to maintain pH 7.0, 6.5, 6.0, or 5.5 for 7 days at each pH, at which time dominant bacterial populations were enumerated. Three representative experiments are shown, using stools from different subjects.
FIG. 3.
Expression of aggR, aafD, and aap from EAEC 042 grown at different pH setpoints in the simulator culture. (A) After equilibration of the system for 72 h, EAEC 042 was inoculated at a final concentration 107 CFU/ml, and incubation was continued for 4 to 18 h. Real-time qRT-PCR was performed for EAEC virulence genes as described in Materials and Methods. Data are reported in arbitrary units of transcript abundance normalized against cat transcript abundance (quantity of target gene/quantity of cat gene). Three representative experiments (E-1, E-2, and E-3) are shown (A). For E-1, samples were only withdrawn 18 h after 042 inoculation. For E-2 and E-3, fermenter samples before EAEC 042 inoculation were included as negative controls. (B) Effect of increasing and decreasing pH on cat, aggR, aafD, and aap expression in the simulator system. The pH setpoint was increased from 5.5 to a maximum of 7.0 and then reduced to 6.0. The system spent at least 24 h at each setpoint. Real- Time qRT-PCR was performed as for Fig. 3A. Data are expressed in arbitrary units of transcript abundance normalized against cat transcript abundance.
FIG. 4.
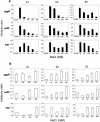
Levels of aggR, aafD, and aap expression in EAEC 042 grown at pH 7.0 (A) or 6.0 (B) in the intestinal simulator. EAEC 042 was grown in the intestinal simulator at the respective pH setpoint, and the expression levels of the three virulence genes were compared directly by real-time qRT-PCR. Data are expressed in arbitrary units of transcript abundance normalized against cat.
FIG. 5.
Effect of the NaCl concentration on EAEC virulence gene expression in the intestinal simulator. (A) EAEC 042 was inoculated once at the beginning of the experiment and grown at different NaCl concentrations for 7 days in the simulator system including fecal flora. (B) EAEC 042 was inoculated anaerobically in simulator medium under continuous flow at various NaCl concentrations, in the absence of fecal commensals. After overnight growth at the osmotic setpoint, total RNA was isolated and cDNA was synthesized and analyzed for cat, aggR, aafD, and aap expression by real-time qRT-PCR. Data represent transcript abundance from three separate experiments normalized against cat transcript.
FIG. 6.
Expression of EAEC virulence genes at various pH setpoints in the absence of commensal flora. Experiments were performed as for Fig. 3, except that no fecal bacteria were added to the system.
FIG. 7.
Effects of commensal flora on aggR expression. EAEC 042 was anaerobically cocultured (A) with specific strains of intestinal bacteria in fermenter medium at pH 6.0 or cultured in media preconditioned (B) by growth of the same bacterial strains. After 18 h of growth in either system, the level of aggR expression was determined by real-time qRT-PCR. Data are expressed as fold increases or decreases of aggR transcript in coculture or preconditioned medium, normalized against aggR abundance in pure cultures of 042 or medium preconditioned by growth of 042.
Similar articles
- Autoactivation of the AggR regulator of enteroaggregative Escherichia coli in vitro and in vivo.
Morin N, Tirling C, Ivison SM, Kaur AP, Nataro JP, Steiner TS. Morin N, et al. FEMS Immunol Med Microbiol. 2010 Apr;58(3):344-55. doi: 10.1111/j.1574-695X.2010.00645.x. Epub 2010 Jan 28. FEMS Immunol Med Microbiol. 2010. PMID: 20132305 - Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli.
Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Dudley EG, et al. Mol Microbiol. 2006 Sep;61(5):1267-82. doi: 10.1111/j.1365-2958.2006.05281.x. Mol Microbiol. 2006. PMID: 16925558 - Organization and architecture of AggR-dependent promoters from enteroaggregative Escherichia coli.
Yasir M, Icke C, Abdelwahab R, Haycocks JR, Godfrey RE, Sazinas P, Pallen MJ, Henderson IR, Busby SJW, Browning DF. Yasir M, et al. Mol Microbiol. 2019 Feb;111(2):534-551. doi: 10.1111/mmi.14172. Epub 2018 Dec 18. Mol Microbiol. 2019. PMID: 30485564 Free PMC article. - Enteroaggregative Escherichia coli pathogenesis.
Nataro JP. Nataro JP. Curr Opin Gastroenterol. 2005 Jan;21(1):4-8. Curr Opin Gastroenterol. 2005. PMID: 15687877 Review. - Pathogenesis of enteroaggregative Escherichia coli infection.
Harrington SM, Dudley EG, Nataro JP. Harrington SM, et al. FEMS Microbiol Lett. 2006 Jan;254(1):12-8. doi: 10.1111/j.1574-6968.2005.00005.x. FEMS Microbiol Lett. 2006. PMID: 16451173 Review.
Cited by
- Investigating Oral Microbiome Profiles in Children with Cleft Lip and Palate for Prognosis of Alveolar Bone Grafting.
Liu L, Zhang Q, Lin J, Ma L, Zhou Z, He X, Jia Y, Chen F. Liu L, et al. PLoS One. 2016 May 18;11(5):e0155683. doi: 10.1371/journal.pone.0155683. eCollection 2016. PLoS One. 2016. PMID: 27191390 Free PMC article. - Characterization of the AggR regulon in enteroaggregative Escherichia coli.
Morin N, Santiago AE, Ernst RK, Guillot SJ, Nataro JP. Morin N, et al. Infect Immun. 2013 Jan;81(1):122-32. doi: 10.1128/IAI.00676-12. Epub 2012 Oct 22. Infect Immun. 2013. PMID: 23090962 Free PMC article. - Nitazoxanide inhibits biofilm production and hemagglutination by enteroaggregative Escherichia coli strains by blocking assembly of AafA fimbriae.
Shamir ER, Warthan M, Brown SP, Nataro JP, Guerrant RL, Hoffman PS. Shamir ER, et al. Antimicrob Agents Chemother. 2010 Apr;54(4):1526-33. doi: 10.1128/AAC.01279-09. Epub 2010 Jan 19. Antimicrob Agents Chemother. 2010. PMID: 20086145 Free PMC article. - Bacterial cell-to-cell signaling in the gastrointestinal tract.
Kaper JB, Sperandio V. Kaper JB, et al. Infect Immun. 2005 Jun;73(6):3197-209. doi: 10.1128/IAI.73.6.3197-3209.2005. Infect Immun. 2005. PMID: 15908344 Free PMC article. Review. No abstract available. - Anaerobic conditions promote expression of Sfp fimbriae and adherence of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM to human intestinal epithelial cells.
Müsken A, Bielaszewska M, Greune L, Schweppe CH, Müthing J, Schmidt H, Schmidt MA, Karch H, Zhang W. Müsken A, et al. Appl Environ Microbiol. 2008 Feb;74(4):1087-93. doi: 10.1128/AEM.02496-07. Epub 2007 Dec 14. Appl Environ Microbiol. 2008. PMID: 18083855 Free PMC article.
References
- Alander, M., I. De Smet, L. Nollet, W. Verstraete, A. Von Wright, and T. Mattila-Sandholm. 1999. The effect of probiotic strains on the microbiota of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME). Int. J. Food Microbiol. 46:71-79. - PubMed
- Berg, Rodney, D. 1996. The indigenous gastrointestinal microflora. Trends Microbiol. 4:430-435. - PubMed
- De Boever, P., B. Deplancke, and W. Verstraete. 2000. Fermentation by gut microbiota cultured in a simulator of the human intestinal microbial ecosystem is improved by supplementing a soygerm powder. J. Nutr. 130:2599-2606. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials