DEQOR: a web-based tool for the design and quality control of siRNAs - PubMed (original) (raw)
. 2004 Jul 1;32(Web Server issue):W113-20.
doi: 10.1093/nar/gkh408.
Affiliations
- PMID: 15215362
- PMCID: PMC441546
- DOI: 10.1093/nar/gkh408
DEQOR: a web-based tool for the design and quality control of siRNAs
Andreas Henschel et al. Nucleic Acids Res. 2004.
Abstract
RNA interference (RNAi) is a powerful tool for inhibiting the expression of a gene by mediating the degradation of the corresponding mRNA. The basis of this gene-specific inhibition is small, double-stranded RNAs (dsRNAs), also referred to as small interfering RNAs (siRNAs), that correspond in sequence to a part of the exon sequence of a silenced gene. The selection of siRNAs for a target gene is a crucial step in siRNA-mediated gene silencing. According to present knowledge, siRNAs must fulfill certain properties including sequence length, GC-content and nucleotide composition. Furthermore, the cross-silencing capability of dsRNAs for other genes must be evaluated. When designing siRNAs for chemical synthesis, most of these criteria are achievable by simple sequence analysis of target mRNAs, and the specificity can be evaluated by a single BLAST search against the transcriptome of the studied organism. A different method for raising siRNAs has, however, emerged which uses enzymatic digestion to hydrolyze long pieces of dsRNA into shorter molecules. These endoribonuclease-prepared siRNAs (esiRNAs or 'diced' RNAs) are less variable in their silencing capabilities and circumvent the laborious process of sequence selection for RNAi due to a broader range of products. Though powerful, this method might be more susceptible to cross-silencing genes other than the target itself. We have developed a web-based tool that facilitates the design and quality control of siRNAs for RNAi. The program, DEQOR, uses a scoring system based on state-of-the-art parameters for siRNA design to evaluate the inhibitory potency of siRNAs. DEQOR, therefore, can help to predict (i) regions in a gene that show high silencing capacity based on the base pair composition and (ii) siRNAs with high silencing potential for chemical synthesis. In addition, each siRNA arising from the input query is evaluated for possible cross-silencing activities by performing BLAST searches against the transcriptome or genome of a selected organism. DEQOR can therefore predict the probability that an mRNA fragment will cross-react with other genes in the cell and helps researchers to design experiments to test the specificity of esiRNAs or chemically designed siRNAs. DEQOR is freely available at http://cluster-1.mpi-cbg.de/Deqor/deqor.html.
Figures
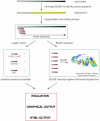
Figure 1
Workflow of a DEQOR analysis. First, the input query is submitted to a regular BLASTN search against the selected transcriptome to identify the origin of the input query. Second, the input query is digested in silico into small sequence pieces (between 16 and 25 nt), mimicking siRNAs. Third, each in silico siRNA is submitted to a BLASTN search against the selected transcriptome. Each siRNA is further analyzed for its sequence properties (see text), and disadvantageous sequence features are penalized. Finally, the siRNAs resulting from an input sequence are sorted according to their quality score and their cross-silencing capacities.
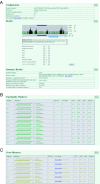
Figure 2
Typical output of a DEQOR analysis. The output window is divided into four major parts. (A) A summary section displays the chosen configuration of the DEQOR run, an interactive graphical display of the quality scores of siRNAs along the input sequence, the putative origin of the input sequence and its length and finally a statistical analysis of the overall quality and cross-silencing capacities. (B) The 10 top quality windows are listed, showing their position in the sequence (‘Window’), the sequence of the siRNA (‘Sequence’, with anti-sense strand in boldface), the number of perfect matches (‘Perf. M.’), matches with one mismatch (‘w/1 Mism.’, the percentage of GC–content (‘GC%’), the asymmetric features of the siRNA (‘ASP’; anti-sense preference, with the options ‘yes’, ‘no’ and ‘sym.’), the occurrence of more than three consecutive identical nucleotides in the siRNA sequence (‘Poly’) and finally the quality score of the siRNA (‘Quality’). (C) Windows that are potential cross-silencers are shown. The columns are identical to (B). The accession numbers (NCBI/ENSEMBL) of genes that are targets for cross-silencing activities are listed and linked to their source database at the NCBI/ENSEMBL. Mouse Dip13 α (NM_145221) was used for this DEQOR analysis.
Figure 3
Statistical analysis of esiRNAs from 3500 randomly selected human genes. (A) The % of siRNAs per gene that have a quality score better than 5 (closed circles) and the % siRNAs that meet all standard quality criteria (open circles) were plotted against the % genes. (B) The average quality scores of each input sequence without (dark gray) and with (light gray) cross-silencing capabilities were related to the % genes in the dataset. Quality scores were divided into four categories: (i) below 5; (ii) between 5 and 10; (iii) between 10 and 20; and (iv) above 20. (C) The % high-quality cross-silencers is plotted against the % genes in the dataset. Only 8.49% of genes have one or more cross-silencers with a quality score below 5 (see text for further discussion).
Similar articles
- dsCheck: highly sensitive off-target search software for double-stranded RNA-mediated RNA interference.
Naito Y, Yamada T, Matsumiya T, Ui-Tei K, Saigo K, Morishita S. Naito Y, et al. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W589-91. doi: 10.1093/nar/gki419. Nucleic Acids Res. 2005. PMID: 15980542 Free PMC article. - Computational siRNA design considering alternative splicing.
Kim YJ. Kim YJ. Methods Mol Biol. 2010;623:81-92. doi: 10.1007/978-1-60761-588-0_5. Methods Mol Biol. 2010. PMID: 20217545 - Using OligoWalk to identify efficient siRNA sequences.
Mathews DH. Mathews DH. Methods Mol Biol. 2010;629:109-21. doi: 10.1007/978-1-60761-657-3_8. Methods Mol Biol. 2010. PMID: 20387146 - On the art of identifying effective and specific siRNAs.
Pei Y, Tuschl T. Pei Y, et al. Nat Methods. 2006 Sep;3(9):670-6. doi: 10.1038/nmeth911. Nat Methods. 2006. PMID: 16929310 Review. - RNA interference--significance and applications.
Stanisławska J, Olszewski WL. Stanisławska J, et al. Arch Immunol Ther Exp (Warsz). 2005 Jan-Feb;53(1):39-46. Arch Immunol Ther Exp (Warsz). 2005. PMID: 15761375 Review.
Cited by
- Integrative and perturbation-based analysis of the transcriptional dynamics of TGFβ/BMP system components in transition from embryonic stem cells to neural progenitors.
Dries R, Stryjewska A, Coddens K, Okawa S, Notelaers T, Birkhoff J, Dekker M, Verfaillie CM, Del Sol A, Mulugeta E, Conidi A, Grosveld FG, Huylebroeck D. Dries R, et al. Stem Cells. 2020 Feb;38(2):202-217. doi: 10.1002/stem.3111. Epub 2019 Dec 13. Stem Cells. 2020. PMID: 31675135 Free PMC article. - Comparing artificial neural networks, general linear models and support vector machines in building predictive models for small interfering RNAs.
McQuisten KA, Peek AS. McQuisten KA, et al. PLoS One. 2009 Oct 22;4(10):e7522. doi: 10.1371/journal.pone.0007522. PLoS One. 2009. PMID: 19847297 Free PMC article. - Efficient prediction of siRNAs with siRNArules 1.0: an open-source JAVA approach to siRNA algorithms.
Holen T. Holen T. RNA. 2006 Sep;12(9):1620-5. doi: 10.1261/rna.81006. Epub 2006 Jul 26. RNA. 2006. PMID: 16870995 Free PMC article. - The major yolk protein vitellogenin interferes with the anti-plasmodium response in the malaria mosquito Anopheles gambiae.
Rono MK, Whitten MM, Oulad-Abdelghani M, Levashina EA, Marois E. Rono MK, et al. PLoS Biol. 2010 Jul 20;8(7):e1000434. doi: 10.1371/journal.pbio.1000434. PLoS Biol. 2010. PMID: 20652016 Free PMC article. - RNA interference for improving the outcome of islet transplantation.
Li F, Mahato RI. Li F, et al. Adv Drug Deliv Rev. 2011 Jan-Feb;63(1-2):47-68. doi: 10.1016/j.addr.2010.11.003. Epub 2010 Dec 13. Adv Drug Deliv Rev. 2011. PMID: 21156190 Free PMC article. Review.
References
- Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. - PubMed
- Denli A.M. and Hannon,G.J. (2003) RNAi: an ever-growing puzzle. Trends Biochem Sci., 28, 196–201. - PubMed
- Cogoni C. and Macino,G. (2000) Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev., 10, 638–643. - PubMed
- Sharp P.A. and Zamore,P.D. (2000) Molecular biology. RNA interference. Science, 287, 2431–2433. - PubMed
- Sharp P.A. (2001) RNA interference—2001. Genes Dev., 15, 485–490. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous