Riboswitch finder--a tool for identification of riboswitch RNAs - PubMed (original) (raw)
. 2004 Jul 1;32(Web Server issue):W154-9.
doi: 10.1093/nar/gkh352.
Affiliations
- PMID: 15215370
- PMCID: PMC441490
- DOI: 10.1093/nar/gkh352
Riboswitch finder--a tool for identification of riboswitch RNAs
Peter Bengert et al. Nucleic Acids Res. 2004.
Abstract
We describe a dedicated RNA motif search program and web server to identify RNA riboswitches. The Riboswitch finder analyses a given sequence using the web interface, checks specific sequence elements and secondary structure, calculates and displays the energy folding of the RNA structure and runs a number of tests including this information to determine whether high-sensitivity riboswitch motifs (or variants) according to the Bacillus subtilis type are present in the given RNA sequence. Batch-mode determination (all sequences input at once and separated by FASTA format) is also possible. The program has been implemented and is available both as local software for in-house installation and as a web server at http://www.biozentrum.uni-wuerzburg.de/bioinformatik/Riboswitch/.
Figures
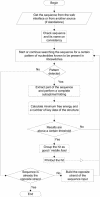
Figure 1
Program flow. After getting the sequence from the web interface, a pattern search including secondary structure and RNA folding including energy values are done. All results are collected and it is scored whether a good, middle or low/tentative riboswitch is present.
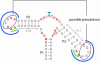
Figure 2
Different consensus options. The general structure of a high-sensitivity guanine riboswitch is shown. The nucleotides marked in red and bold define the strict consensus. No mismatches are allowed here. The option ‘general consensus’ allows mismatches (marked in green) in the loop regions of P2 and P3 that are not part of the possible pseudoknot and one mismatch in the connection between P2 and P3. Option ‘loose consensus’ allows mismatches (marked in blue) in both loops P2 and P3, no matter whether the nucleotides are part of the possible pseudoknot or not.
Figure 3
Web interface for the riboswitch finder.
Figure 4
Result of a search with the riboswitch finder (example file STPY1). Folding results, energy, and base pairings are shown, giving the exact region where the riboswitch starts and a graphical display at the bottom of the web page. In the example the program identifies from the sequence file a known riboswitch in the 5′-UTR of xanthine phosphoribosyltransferase in S.pyogenes. Three possible overlapping patterns describing this riboswitch element, each compatible with the consensus structure, are printed out.
Similar articles
- A software tool-box for analysis of regulatory RNA elements.
Bengert P, Dandekar T. Bengert P, et al. Nucleic Acids Res. 2003 Jul 1;31(13):3441-5. doi: 10.1093/nar/gkg568. Nucleic Acids Res. 2003. PMID: 12824342 Free PMC article. - RegRNA: an integrated web server for identifying regulatory RNA motifs and elements.
Huang HY, Chien CH, Jen KH, Huang HD. Huang HY, et al. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W429-34. doi: 10.1093/nar/gkl333. Nucleic Acids Res. 2006. PMID: 16845041 Free PMC article. - RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements.
Abreu-Goodger C, Merino E. Abreu-Goodger C, et al. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W690-2. doi: 10.1093/nar/gki445. Nucleic Acids Res. 2005. PMID: 15980564 Free PMC article. - [The adenine riboswitch: a new gene regulation mechanism].
Lemay JF, Lafontaine DA. Lemay JF, et al. Med Sci (Paris). 2006 Dec;22(12):1053-9. doi: 10.1051/medsci/200622121053. Med Sci (Paris). 2006. PMID: 17156726 Review. French. - Determination of riboswitch structures: light at the end of the tunnel?
Serganov A. Serganov A. RNA Biol. 2010 Jan-Feb;7(1):98-103. doi: 10.4161/rna.7.1.10756. Epub 2010 Jan 25. RNA Biol. 2010. PMID: 20061809 Review.
Cited by
- A novel riboswitch classification based on imbalanced sequences achieved by machine learning.
Beyene SS, Ling T, Ristevski B, Chen M. Beyene SS, et al. PLoS Comput Biol. 2020 Jul 20;16(7):e1007760. doi: 10.1371/journal.pcbi.1007760. eCollection 2020 Jul. PLoS Comput Biol. 2020. PMID: 32687488 Free PMC article. - Guanine riboswitch variants from Mesoplasma florum selectively recognize 2'-deoxyguanosine.
Kim JN, Roth A, Breaker RR. Kim JN, et al. Proc Natl Acad Sci U S A. 2007 Oct 9;104(41):16092-7. doi: 10.1073/pnas.0705884104. Epub 2007 Oct 2. Proc Natl Acad Sci U S A. 2007. PMID: 17911257 Free PMC article. - Adding context to the pneumococcal core genes using bioinformatic analysis of the intergenic pangenome of Streptococcus pneumoniae.
Nielsen FD, Møller-Jensen J, Jørgensen MG. Nielsen FD, et al. Front Bioinform. 2023 Feb 8;3:1074212. doi: 10.3389/fbinf.2023.1074212. eCollection 2023. Front Bioinform. 2023. PMID: 36844929 Free PMC article. - Structural introspection of a putative fluoride transporter in plants.
Banerjee A, Roychoudhury A. Banerjee A, et al. 3 Biotech. 2019 Mar;9(3):103. doi: 10.1007/s13205-019-1629-4. Epub 2019 Feb 22. 3 Biotech. 2019. PMID: 30800614 Free PMC article. - Riboflow: Using Deep Learning to Classify Riboswitches With ∼99% Accuracy.
Premkumar KAR, Bharanikumar R, Palaniappan A. Premkumar KAR, et al. Front Bioeng Biotechnol. 2020 Jul 14;8:808. doi: 10.3389/fbioe.2020.00808. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32760712 Free PMC article.
References
- Stormo G.D. (2003) New tricks for an old dogma: riboswitches as cis-only regulatory systems. Mol. Cell, 11, 1419–1420. - PubMed
- Mandal M., Boese,B., Barrick,J.E., Winkler,W.C. and Breaker,R.R. (2003) Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell, 113, 577–586. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources