RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons - PubMed (original) (raw)
. 2004 Jul 1;32(Web Server issue):W187-90.
doi: 10.1093/nar/gkh393.
Affiliations
- PMID: 15215377
- PMCID: PMC441531
- DOI: 10.1093/nar/gkh393
RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons
William G Fairbrother et al. Nucleic Acids Res. 2004.
Abstract
A typical gene contains two levels of information: a sequence that encodes a particular protein and a host of other signals that are necessary for the correct expression of the transcript. While much attention has been focused on the effects of sequence variation on the amino acid sequence, variations that disrupt gene processing signals can dramatically impact gene function. A variation that disrupts an exonic splicing enhancer (ESE), for example, could cause exon skipping which would result in the exclusion of an entire exon from the mRNA transcript. RESCUE-ESE, a computational approach used in conjunction with experimental validation, previously identified 238 candidate ESE hexamers in human genes. The RESCUE-ESE method has recently been implemented in three additional species: mouse, zebrafish and pufferfish. Here we describe an online ESE analysis tool (http://genes.mit.edu/burgelab/rescue-ese/) that annotates RESCUE-ESE hexamers in vertebrate exons and can be used to predict splicing phenotypes by identifying sequence changes that disrupt or alter predicted ESEs.
Figures
Figure 1
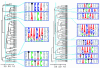
ESE hexamers for the 3′ss and the 5′ss have been clustered into 10 distinct motifs. The RESCUE-ESE protocol identified (A) 103 hexamers from the 5′ss and (B) 198 hexamers from the 3′ss which, when clustered on the basis of sequence similarity (penalizing 1 point for a mismatch or shift), could be aligned to form five ESE motifs for the 5′ss and eight motifs for the 3′ss. The dissimilarity between hexamers (or average dissimilarity between clusters of hexamers) is shown on the bottom scale, where the blue line represents the threshold dissimilarity of 2.7 that was used to define the clusters. The weight matrices that define the ESE motifs are represented as pictograms where the size of each letter is proportional to the nucleotide frequency for each of the 7–10 positions in the ESE motif.
Figure 2
The RESCUE-ESE web server. An exon skipping mutation from the human HPRT gene was analyzed by RESCUE-ESE. (A) Multi FASTA format files containing the wild type and mutant sequences of an A to T mutation at HPRT position 163 were pasted into the input window on the entry page of
http://genes.mit.edu/burgelab/rescue-ese
. (B) Species buttons allow the user to select from multiple sets of RESCUE-ESE hexamers. The output displays the input sequence with the ESEs drawn above in yellow. Additional links include ESE references, the original RESCUE-ESE paper and download options for ESE hexamers.
Similar articles
- Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes.
Wang J, Smith PJ, Krainer AR, Zhang MQ. Wang J, et al. Nucleic Acids Res. 2005 Sep 7;33(16):5053-62. doi: 10.1093/nar/gki810. Print 2005. Nucleic Acids Res. 2005. PMID: 16147989 Free PMC article. - ESEfinder: A web resource to identify exonic splicing enhancers.
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. Cartegni L, et al. Nucleic Acids Res. 2003 Jul 1;31(13):3568-71. doi: 10.1093/nar/gkg616. Nucleic Acids Res. 2003. PMID: 12824367 Free PMC article. - Predictive identification of exonic splicing enhancers in human genes.
Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Fairbrother WG, et al. Science. 2002 Aug 9;297(5583):1007-13. doi: 10.1126/science.1073774. Epub 2002 Jul 11. Science. 2002. PMID: 12114529 - Bioinformatics and mutations leading to exon skipping.
Desmet FO, Béroud C. Desmet FO, et al. Methods Mol Biol. 2012;867:17-35. doi: 10.1007/978-1-61779-767-5_2. Methods Mol Biol. 2012. PMID: 22454052 Review. - Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases.
Blencowe BJ. Blencowe BJ. Trends Biochem Sci. 2000 Mar;25(3):106-10. doi: 10.1016/s0968-0004(00)01549-8. Trends Biochem Sci. 2000. PMID: 10694877 Review.
Cited by
- Antisense therapy in neurology.
Lee JJ, Yokota T. Lee JJ, et al. J Pers Med. 2013 Aug 2;3(3):144-76. doi: 10.3390/jpm3030144. J Pers Med. 2013. PMID: 25562650 Free PMC article. Review. - Sequence mutations of the substrate binding pocket of stem cell factor and multidrug resistance protein ABCG2 in renal cell cancer: a possible link to treatment resistance.
Zoernig I, Ziegelmeier C, Lahrmann B, Grabe N, Jäger D, Halama N. Zoernig I, et al. Oncol Rep. 2013 May;29(5):1697-700. doi: 10.3892/or.2013.2324. Epub 2013 Mar 5. Oncol Rep. 2013. PMID: 23467750 Free PMC article. - Cryptic exon activation by disruption of exon splice enhancer: novel mechanism causing 3-methylcrotonyl-CoA carboxylase deficiency.
Stucki M, Suormala T, Fowler B, Valle D, Baumgartner MR. Stucki M, et al. J Biol Chem. 2009 Oct 16;284(42):28953-7. doi: 10.1074/jbc.M109.050674. Epub 2009 Aug 24. J Biol Chem. 2009. PMID: 19706617 Free PMC article. - Exon Skipping in a Dysf-Missense Mutant Mouse Model.
Malcher J, Heidt L, Goyenvalle A, Escobar H, Marg A, Beley C, Benchaouir R, Bader M, Spuler S, García L, Schöwel V. Malcher J, et al. Mol Ther Nucleic Acids. 2018 Dec 7;13:198-207. doi: 10.1016/j.omtn.2018.08.013. Epub 2018 Aug 22. Mol Ther Nucleic Acids. 2018. PMID: 30292141 Free PMC article. - The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons.
Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. Minovitsky S, et al. Nucleic Acids Res. 2005 Feb 3;33(2):714-24. doi: 10.1093/nar/gki210. Print 2005. Nucleic Acids Res. 2005. PMID: 15691898 Free PMC article.
References
- Fairbrother W.G., Yeh,R.F., Sharp,P.A. and Burge,C.B. (2002) Predictive identification of exonic splicing enhancers in human genes. Science, 297, 1007–1013. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources