Labeling of fusion proteins with synthetic fluorophores in live cells - PubMed (original) (raw)
Labeling of fusion proteins with synthetic fluorophores in live cells
Antje Keppler et al. Proc Natl Acad Sci U S A. 2004.
Abstract
A general approach for the sequential labeling of fusion proteins of O(6)-alkylguanine-DNA alkyltransferase (AGT) with different fluorophores in mammalian cells is presented. AGT fusion proteins with different localizations in the cell can be labeled specifically with different fluorophores, and the fluorescence labeling can be used for applications such as multicolor analysis of dynamic processes and fluorescence resonance energy transfer measurements. The facile access to a variety of different AGT substrates as well as the specificity of the labeling reaction should make the approach an important tool to study protein function in live cells.
Figures
Fig. 1.
General mechanism of (A) and substrates used for (B) fluorescence labeling of AGT fusion proteins. (A) Covalent labeling of AGT fusion proteins with BG derivatives. (B) Structures of BG derivatives used for fluorescence labeling of AGT fusion proteins. The maxima in the absorbance (Abs.) and fluorescence emission spectra (Em.) of the substrates after reaction with AGT are listed; the data for BGAF, BGOG, and BGSF are those of the deacetylated molecules.
Fig. 2.
Transiently transfected CHO cells expressing various AGT fusion proteins labeled with fluorescent BG derivatives. (A) AGT-NLS3 labeled with OG using BGOG. (B) AGT-NLS3 labeled with TMR using BGTMR. (C) AGT-NLS3 labeled with SF using BGSF. (D) AGT-β-Gal labeled with fluorescein using BGAF. (E) AGT labeled with fluorescein. (F) EGFP. (G) AGT-CaaX labeled with TMR. (H) AGT-α-tubulin labeled with fluorescein. (I) tsVSVG-AGT labeled with fluorescein. (Scale bars, 10 μm.)
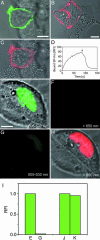
Fig. 3.
Extracellular (A_–_D) and FRET (E_–_I) applications of AGT fusion proteins. (A_–_D) Transiently transfected HEK293 cells expressing AGT-NK1. (A) AGT-NK1 labeled with fluorescein. (B) AGT-NK1 labeled with Cy3 using BGCy3. (C) Binding of SP-rho (50 nM) to the same cell as in A. (D) Time-resolved binding of SP-rho (50 nM) to an HEK293 cell displaying AGT-NK1 and its replacement through the addition of unlabeled ligand (2.5 μM) at the indicated time (black arrow). (E_–_I) Transiently transfected CHO cells expressing AGT-EGFP-NLS3 labeled with SF for FRET detection. (E) Overlay of transmission and fluorescence channel for EGFP before labeling with SF (excitation 488 nm/emission 505–530 nm). (F) Fluorescence channel for SF before labeling with SF (excitation 488 nm/emission > 650 nm). (G) Fluorescence channel for EGFP 60 min after labeling with SF (excitation 488 nm/emission 505–530 nm). (H) Overlay of transmission and fluorescence channel for SF after labeling with SF (excitation 488 nm/emission > 650 nm). (I) Relative fluorescence intensities (RFI) of the EGFP channel before and after labeling with SF. Columns E and G correspond to the fluorescence intensities (RFI) of the EGFP channel of the cell shown in E and G. Columns J and K correspond to the fluorescence intensities (RFI) of the EGFP channel of a cell before and after incubation with SF but previously incubated with BG (20 μM). (Scale bars 10 μm.)
Fig. 4.
Multicolor analysis of tsVSVG-AGT and labeling of AGT-CaaX in cells possessing endogenous AGT. (A_–_C) Sequential labeling of tsVSVG-AGT: Labeling with fluorescein at permissive temperature (34°C) and with SF at nonpermissive temperature (40°C). (A) Overlay of transmission and fluorescence micrographs. (B) Fluorescence channel for fluorescein-labeled tsVSVG-AGT (excitation 488 nm/emission 505–530 nm). (C) Fluorescence channel for SF-labeled tsVSVG-AGT (excitation 543 nm/emission > 650 nm). (D and E) Labeling of AGT-CaaX in HEK293 cells possessing endogenous AGT. (D) Fluorescence channel after labeling with fluorescein (excitation 488 nm/emission 505–530 nm). (E) Transmission channel of same cells as in G. (Scale bar, 10 μm.)
Similar articles
- Engineering substrate specificity of O6-alkylguanine-DNA alkyltransferase for specific protein labeling in living cells.
Juillerat A, Heinis C, Sielaff I, Barnikow J, Jaccard H, Kunz B, Terskikh A, Johnsson K. Juillerat A, et al. Chembiochem. 2005 Jul;6(7):1263-9. doi: 10.1002/cbic.200400431. Chembiochem. 2005. PMID: 15934048 - Fluorophores for live cell imaging of AGT fusion proteins across the visible spectrum.
Keppler A, Arrivoli C, Sironi L, Ellenberg J. Keppler A, et al. Biotechniques. 2006 Aug;41(2):167-70, 172, 174-5. doi: 10.2144/000112216. Biotechniques. 2006. PMID: 16925018 - Site-specific, covalent labeling of recombinant antibody fragments via fusion to an engineered version of 6-O-alkylguanine DNA alkyltransferase.
Kampmeier F, Ribbert M, Nachreiner T, Dembski S, Beaufils F, Brecht A, Barth S. Kampmeier F, et al. Bioconjug Chem. 2009 May 20;20(5):1010-5. doi: 10.1021/bc9000257. Bioconjug Chem. 2009. PMID: 19388673 - Insight into the cooperative DNA binding of the O⁶-alkylguanine DNA alkyltransferase.
Tessmer I, Fried MG. Tessmer I, et al. DNA Repair (Amst). 2014 Aug;20:14-22. doi: 10.1016/j.dnarep.2014.01.006. Epub 2014 Feb 16. DNA Repair (Amst). 2014. PMID: 24553127 Free PMC article. Review. - The Evolution of SNAP-Tag Labels.
Dreyer R, Pfukwa R, Barth S, Hunter R, Klumperman B. Dreyer R, et al. Biomacromolecules. 2023 Feb 13;24(2):517-530. doi: 10.1021/acs.biomac.2c01238. Epub 2023 Jan 6. Biomacromolecules. 2023. PMID: 36607253 Review.
Cited by
- Aged insulin granules display reduced microtubule-dependent mobility and are disposed within actin-positive multigranular bodies.
Hoboth P, Müller A, Ivanova A, Mziaut H, Dehghany J, Sönmez A, Lachnit M, Meyer-Hermann M, Kalaidzidis Y, Solimena M. Hoboth P, et al. Proc Natl Acad Sci U S A. 2015 Feb 17;112(7):E667-76. doi: 10.1073/pnas.1409542112. Epub 2015 Feb 2. Proc Natl Acad Sci U S A. 2015. PMID: 25646459 Free PMC article. - Emissive Alkylated Guanine Analogs as Probes for Monitoring O 6-Alkylguanine-DNA-transferase Activity.
Steinbuch KB, Bucardo M, Tor Y. Steinbuch KB, et al. ACS Omega. 2024 Aug 17;9(34):36778-36786. doi: 10.1021/acsomega.4c05700. eCollection 2024 Aug 27. ACS Omega. 2024. PMID: 39220506 Free PMC article. - Simple and efficient delivery of cell-impermeable organic fluorescent probes into live cells for live-cell superresolution imaging.
Zhang M, Li M, Zhang W, Han Y, Zhang YH. Zhang M, et al. Light Sci Appl. 2019 Aug 14;8:73. doi: 10.1038/s41377-019-0188-0. eCollection 2019. Light Sci Appl. 2019. PMID: 31666945 Free PMC article. - Development of SNAP-tag fluorogenic probes for wash-free fluorescence imaging.
Sun X, Zhang A, Baker B, Sun L, Howard A, Buswell J, Maurel D, Masharina A, Johnsson K, Noren CJ, Xu MQ, Corrêa IR Jr. Sun X, et al. Chembiochem. 2011 Sep 19;12(14):2217-26. doi: 10.1002/cbic.201100173. Epub 2011 Jul 26. Chembiochem. 2011. PMID: 21793150 Free PMC article. - Two-color STED microscopy in living cells.
Pellett PA, Sun X, Gould TJ, Rothman JE, Xu MQ, Corrêa IR Jr, Bewersdorf J. Pellett PA, et al. Biomed Opt Express. 2011 Aug 1;2(8):2364-71. doi: 10.1364/BOE.2.002364. Epub 2011 Jul 22. Biomed Opt Express. 2011. PMID: 21833373 Free PMC article.
References
- Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. (2002) Nat. Rev. Mol. Cell Biol. 3, 906-918. - PubMed
- Lippincott-Schwartz, J. & Patterson, G. H. (2003) Science 300, 87-91. - PubMed
- Miyawaki, A., Sawano, A. & Kogure, T. (2003) Nat. Cell Biol. 5, Suppl., S1-S7. - PubMed
- Lippincott-Schwartz, J., Altan-Bonnet, N. & Patterson, G. H. (2003) Nat. Cell Biol. 5, Suppl., S7-S14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous