The gamma2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ - PubMed (original) (raw)
Comparative Study
The gamma2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ
Cheryl A Keller et al. J Neurosci. 2004.
Abstract
The neurotransmitter GABA activates heteropentameric GABA(A) receptors, which are composed mostly of alpha, beta, and gamma2 subunits. Regulated membrane trafficking and subcellular targeting of GABA(A) receptors is important for determining the efficacy of GABAergic inhibitory function. Of special interest is the gamma2 subunit, which is mostly dispensable for assembly and membrane insertion of functional receptors but essential for accumulation of GABA(A) receptors at synapses. In a search for novel receptor trafficking proteins, we have used the SOS-recruitment system and isolated a Golgi-specific DHHC zinc finger protein (GODZ) as a novel gamma2 subunit-interacting protein. GODZ is a member of the superfamily of DHHC cysteine-rich domain (DHHC-CRD) polytopic membrane proteins shown recently in yeast to represent palmitoyltransferases. GODZ mRNA is found in many tissues; however, in brain the protein is detected in neurons only and highly concentrated and asymmetrically distributed in the Golgi complex. GODZ interacts with a cysteine-rich 14-amino acid domain conserved specifically in the large cytoplasmic loop of gamma1-3 subunits but not in other GABA(A) receptor subunits. Coexpression of GODZ and GABA(A) receptors in heterologous cells results in palmitoylation of the gamma2 subunit in a cytoplasmic loop domain-dependent manner. Neuronal GABA(A) receptors are similarly palmitoylated. Thus, GODZ-mediated palmitoylation represents a novel posttranslational modification that is selective for gamma subunit-containing GABA(A) receptor subtypes, a mechanism that is likely to be important for regulated trafficking of these receptors in the secretory pathway.
Figures
Figure 2.
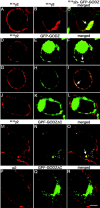
GFP-tagged GODZ partly colocalizes and interacts with γ2 subunit-containing GABAA receptors in transfected HEK 293T cells. A-C, HEK 293T cells were transfected with the (9E10)γ2 subunit alone (A) or together with GFP-GODZ (B, C) and fixed and permeabilized. The γ2 subunit was stained with mAb 9E10 and secondary antibody (red); colocalization with GFP-GODZ (green) is shown in yellow. Note the reduced expression of the γ2 subunit in the plasma membrane and the increased intracellular localization of the γ2 subunit after expression of GFP-GODZ. D-L, HEK 293T cells were cotransfected with α2β3 (9E10)γ2 receptors and GFP-GODZ and processed as above (D-I) or incubated while alive and the receptors aggregated with mAb 9E10 and secondary antibody (J-L). Theγ2 subunit visualized with mAb 9E10 (D-L) is shown in red, GFP-GODZ (E, F, H, I, K, L) is shown in green, and the merged panels (F, I) illustrate colocalization (yellow) or the lack thereof (L). Note the variable subcellular localization of GFP-GODZ. M-R, HEK 293T cells were cotransfected with GFP-GODZΔC and either α2β3 (9E10)γ2 (M-O) or α2β3 receptors (P-R). The cells were fixed, permeabilized, and processed for immunofluorescence using mAb 9E10 or anti-α2 antiserum to visualize the γ2 (M, O) and α2 subunit (P, R), respectively, in red. GFP-GODZΔC is shown in green. Note the localization of GFP-GODZΔC to intracellular compartments (N, Q), accompanied by intracellular accumulation of the γ2 subunit (M, O). In contrast, no such intracellular localization is seen for the α2 subunit when α2β 3 subunits are cotransfected with GFP-GODZΔC in the absence of the γ2 subunit (P-R). Images represent single optical sections. Arrows indicate colocalization. Scale bar, 5 μm.
Figure 1.
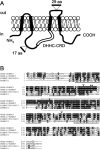
Proposed transmembrane topology of GODZ and deduced amino acid sequence of GODZ with other family members. A, Transmembrane topology of GODZ as predicted by TMpred (see Materials and Methods). Putative membrane-spanning regions (black boxes), the DHHCCRD domain (open box), and the positions of alternative exons (see Results) are indicated. B, Alignment of mouse GODZ/ZDHHC3, SERZ-β/ZDHHC7, RIKEN clone 1700030J15, and ZDHHC21. Identical and similar amino acids are shaded in black and gray, respectively. Membrane-spanning regions are underlined, and the DHHC-CRD domain is boxed with a dashed line. *, Putative PKC phosphorylation site; **, putative glycosylation site; ▿, positions of alternative exons.
Figure 3.
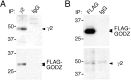
Theγ2 subunit and GODZ from a complex in transfected HEK 293T cells. To directly test whether GABAA receptors and GODZ can exist in a complex in mammalian cells, we immunoprecipitated the γ2 subunit from extracts of HEK 293T cells cotransfected with vectors encoding theγ2 subunit and FLAG-GODZ. A, Western blot analyses of γ2 subunit immunoprecipitates revealed efficient copurification of GODZ. B, Conversely, immunopurification of FLAG-GODZ from the same extracts revealed coimmunoprecipitation of the γ2 subunit, confirming that the two proteins can exist in a complex when overexpressed in HEK 293T cells. Note that the weak unspecific band seen in both the FLAG immunoprecipitate and the IgG control lane (B, bottom panel) runs at 50 kDa just above the γ2 subunit. Results shown are representative of three (A) and five (B) successful experiments, respectively.
Figure 4.
Characterization of γ2 subunit and GODZ interaction domains. A, Various GABAA receptor subunit constructs, each of which contains a portion of its major intracellular loop, were tested as baits for interaction with full-length GODZ and ZDHHC7, respectively. The GODZ interaction domain within the γ2 subunit, and homologous regions within the γ1 and γ3 subunits, are boxed. Note that although the amino acid sequences in the bait construct are 100% conserved between rat and mouse cDNAs in the case of the γ2 and γ3 subunits, the γ1 bait sequence is representative of the rat γ1 subunit only. B, Deletion constructs of the γ2 intracellular loop were used as baits in the yeast two-hybrid system to map the GODZ binding domain. All constructs were tested in combination with full-length GODZ as a prey. The GABARAP binding domain in the γ2 subunit sequence (Wang et al., 1999) is shown to be located C terminal of the GODZ minimal binding domain. C, Various truncations of GODZ cDNA were tested as prey in the yeast two-hybrid system to determine the γ2 interaction domain. All constructs were tested in combination with the original γ2 subunit bait (amino acids 361-404 of γ2S). The original GODZ partial cDNA isolated in the yeast two-hybrid screen is indicated with an asterisk. The strength of interaction is indicated by a semiquantitative scale of colony growth with ++, +, and - indicating strong, weak, and no growth, respectively. The minimal binding domains in the γ2 cytoplasmic loop for GODZ and GABARAP (B), the putative transmembrane regions (M1-M4), and the DHHC-CRD domain of GODZ (C) are indicated.
Figure 5.
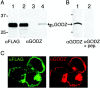
GODZ antiserum recognizes recombinant and native GODZ in vitro and in vivo. A, Immunoblot of extracts prepared from HEK 293T cells transfected with FLAG-GODZ (lanes 1, 2), untransfected HEK 293T cells (lane 3), and brain membranes (lane 4) were visualized using αFLAG (lane 1) or αGODZ (lanes 2-4), respectively. The arrows at ∼31 kDa indicate the mobility of endogenous GODZ and FLAG-GODZ. B, Immunoblot of extracts prepared from brain membranes (lanes 1, 2) developed with either GODZ antiserum (lane 1) or GODZ antiserum that was preadsorbed with the immunizing peptide antigen (lane 2). C, FLAG-GODZ was transfected into HEK 293T cells and stained with an antibody directed against the FLAG epitope tag (green) or with the GODZ antiserum (red). Results shown are representative of at least three similar experiments each.
Figure 6.
GODZ is specifically expressed in neurons and localized to the periphery of the Golgi apparatus. A, Spatial expression of GODZ mRNA in brain was addressed by in situ hybridization of parasagittal brain sections using radiolabeled probes directed to the 3′ untranslated region of the GODZ mRNA. GODZ transcripts are broadly distributed throughout all major brain areas but most concentrated in structures rich in neural cell bodies. B, Confocal image of a rat brain section showing a cerebellar neuron (deep nuclei) double labeled for GODZ (red) and gephyrin (green). C-E, Primary cultured cortical neuron double labeled for GODZ (C, red) and Golgi 58 kDa (D, green). Colocalization is shown in yellow (E). Note that immunoreactivity for Golgi 58 kDa appears not entirely limited to the Golgi complex. F, Pre-embedding immunocytochemistry of GODZ in the hippocampal pyramidal layer. The immunoreaction end product was silver intensified and gold toned and is detected selectively at the periphery of the Golgi complex. Go, Golgi; syn, symmetric synapse. Scale bars: B-E, 5 μm; F, 0.25 μm.
Figure 7.
Theγ2 subunit cytoplasmic loop is a substrate for palmitoylation by GODZ. A-C, Recombinant (9E10)γ2 subunit and FLAG-GODZ were cotransfected and expressed in HEK 293T cells, metabolically labeled with [3H]palmitic acid for 4-5 hr, immunoprecipitated with anti-γ2 antiserum or IgG control serum, and subjected to SDS-PAGE and fluorography (A), Western blot analysis using mAb 9E10 (B), or hydroxylamine treatment before fluorography (C). D, Recombinant α1β(9E10)γ2 receptors together with FLAG-GODZ (lanes 1, 3) or alone (lanes 2, 4) were cotransfected and expressed in HEK 293T cells, metabolically labeled with [3H]palmitic acid for 4-5 hr, immunoprecipitated with anti-γ2 antiserum or IgG control serum, subjected to SDS-PAGE and fluorography (top panel), or visualized by Western blot with mAb 9E10 (bottom panel). E, F, Recombinant (9E10)γ2 and either GFP-GODZ (lane 1) or FLAG-GODZ (lane 2) were cotransfected and expressed in HEK 293T cells, metabolically labeled with [3H]palmitic acid for 4-5 hr, and immunoprecipitated with anti-γ2 antiserum. The sample was divided, subjected to SDS-PAGE, and processed for fluorography (E) and Western blot analysis using αGODZ antiserum (F), respectively. The faster-migrating GFP-GODZ species is likely to represent a degradation product of the full-length product. G, FLAG-GODZ was cotransfected with recombinant α1β2 subunits and (9E10)γ2 (lane 1), empty vector (pRK5, lane 2), or a (9E10)γ2/α2 chimeric construct (lane 3; see Materials and Methods) into HEK 293T cells. The cells were metabolically labeled with [3H]palmitic acid, immunoprecipitated with anti-γ2 antiserum, and fractions of the sample were processed for fluorography (top panel) and Western blot (bottom panel) using an anti-myc antiserum, respectively. H, Cultured cortical neurons were metabolically labeled with [3H]palmitic acid. GABAA receptors were immunoprecipitated with anti-α1 antiserum or IgG control serum and processed for fluorography (top panel) and Western blot (bottom panel) using anti-γ2 antiserum as in G. Results shown are representative of two to three similar experiments each.
Similar articles
- GODZ-mediated palmitoylation of GABA(A) receptors is required for normal assembly and function of GABAergic inhibitory synapses.
Fang C, Deng L, Keller CA, Fukata M, Fukata Y, Chen G, Lüscher B. Fang C, et al. J Neurosci. 2006 Dec 6;26(49):12758-68. doi: 10.1523/JNEUROSCI.4214-06.2006. J Neurosci. 2006. PMID: 17151279 Free PMC article. - Dissociation of Golgi-associated DHHC-type Zinc Finger Protein (GODZ)- and Sertoli Cell Gene with a Zinc Finger Domain-β (SERZ-β)-mediated Palmitoylation by Loss of Function Analyses in Knock-out Mice.
Kilpatrick CL, Murakami S, Feng M, Wu X, Lal R, Chen G, Du K, Luscher B. Kilpatrick CL, et al. J Biol Chem. 2016 Dec 30;291(53):27371-27386. doi: 10.1074/jbc.M116.732768. Epub 2016 Nov 14. J Biol Chem. 2016. PMID: 27875292 Free PMC article. - Interaction between GABAA receptor subunit intracellular loops: implications for higher order complex formation.
Nymann-Andersen J, Sawyer GW, Olsen RW. Nymann-Andersen J, et al. J Neurochem. 2002 Dec;83(5):1164-71. doi: 10.1046/j.1471-4159.2002.01222.x. J Neurochem. 2002. PMID: 12437587 - GABAA receptor associated proteins: a key factor regulating GABAA receptor function.
Chen ZW, Olsen RW. Chen ZW, et al. J Neurochem. 2007 Jan;100(2):279-94. doi: 10.1111/j.1471-4159.2006.04206.x. Epub 2006 Nov 2. J Neurochem. 2007. PMID: 17083446 Review. - Protein Palmitoylation by DHHC Protein Family.
Fukata Y, Bredt DS, Fukata M. Fukata Y, et al. In: Kittler JT, Moss SJ, editors. The Dynamic Synapse: Molecular Methods in Ionotropic Receptor Biology. Boca Raton (FL): CRC Press/Taylor & Francis; 2006. Chapter 5. In: Kittler JT, Moss SJ, editors. The Dynamic Synapse: Molecular Methods in Ionotropic Receptor Biology. Boca Raton (FL): CRC Press/Taylor & Francis; 2006. Chapter 5. PMID: 21204476 Free Books & Documents. Review.
Cited by
- S-acylation of Ca2+ transport proteins: molecular basis and functional consequences.
Néré R, Kouba S, Carreras-Sureda A, Demaurex N. Néré R, et al. Biochem Soc Trans. 2024 Feb 28;52(1):407-421. doi: 10.1042/BST20230818. Biochem Soc Trans. 2024. PMID: 38348884 Free PMC article. Review. - The stress-induced cytokine interleukin-6 decreases the inhibition/excitation ratio in the rat temporal cortex via trans-signaling.
Garcia-Oscos F, Salgado H, Hall S, Thomas F, Farmer GE, Bermeo J, Galindo LC, Ramirez RD, D'Mello S, Rose-John S, Atzori M. Garcia-Oscos F, et al. Biol Psychiatry. 2012 Apr 1;71(7):574-82. doi: 10.1016/j.biopsych.2011.11.018. Epub 2011 Dec 22. Biol Psychiatry. 2012. PMID: 22196984 Free PMC article. - Repositioning Lomitapide to block ZDHHC5-dependant palmitoylation on SSTR5 leads to anti-proliferation effect in preclinical pancreatic cancer models.
Wang Y, Zhang S, He H, Luo H, Xia Y, Jiang Y, Jiang J, Sun L. Wang Y, et al. Cell Death Discov. 2023 Feb 11;9(1):60. doi: 10.1038/s41420-023-01359-4. Cell Death Discov. 2023. PMID: 36774350 Free PMC article. - Palmitoylation-dependent plasma membrane transport but lipid raft-independent signaling by linker for activation of T cells.
Hundt M, Harada Y, De Giorgio L, Tanimura N, Zhang W, Altman A. Hundt M, et al. J Immunol. 2009 Aug 1;183(3):1685-94. doi: 10.4049/jimmunol.0803921. Epub 2009 Jul 10. J Immunol. 2009. PMID: 19592663 Free PMC article. - Subcellular Golgi localization of stathmin family proteins is promoted by a specific set of DHHC palmitoyl transferases.
Levy AD, Devignot V, Fukata Y, Fukata M, Sobel A, Chauvin S. Levy AD, et al. Mol Biol Cell. 2011 Jun 1;22(11):1930-42. doi: 10.1091/mbc.E10-10-0824. Epub 2011 Apr 6. Mol Biol Cell. 2011. PMID: 21471001 Free PMC article.
References
- Aronheim A, Karin M (2000) Analysis and identification of protein-protein interactions using protein recruitment systems. Methods Enzymol 328: 47-59. - PubMed
- Benke D, Honer M, Michel C, Mohler H (1996) GABAA receptor subtypes differentiated by their γ-subunit variants: prevalence, pharmacology and subunit architecture. Neuropharmacology 35: 1413-1423. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH062391-05/MH/NIMH NIH HHS/United States
- NS11070/NS/NINDS NIH HHS/United States
- R01 MH062391-02/MH/NIMH NIH HHS/United States
- R01 MH060989/MH/NIMH NIH HHS/United States
- R01 MH062391/MH/NIMH NIH HHS/United States
- MH62391/MH/NIMH NIH HHS/United States
- R56 MH062391/MH/NIMH NIH HHS/United States
- R01 MH062391-04/MH/NIMH NIH HHS/United States
- R01 MH062391-03/MH/NIMH NIH HHS/United States
- R01 MH062391-01A2/MH/NIMH NIH HHS/United States
- F32 NS011070/NS/NINDS NIH HHS/United States
- R01 MH060989-01A2/MH/NIMH NIH HHS/United States
- R01 MH060989-02/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases