The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation - PubMed (original) (raw)
Comparative Study
. 2004 Jul 15;18(14):1725-36.
doi: 10.1101/gad.303504. Epub 2004 Jul 1.
Anja Beckers, Karin Schuster-Gossler, Maria N Pavlova, Hannelore Burkhardt, Heiko Lickert, Janet Rossant, Richard Reinhardt, Leonard C Schalkwyk, Ines Müller, Bernhard G Herrmann, Marcelo Ceolin, Rolando Rivera-Pomar, Achim Gossler
Affiliations
- PMID: 15231714
- PMCID: PMC478193
- DOI: 10.1101/gad.303504
Comparative Study
The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation
Hanaa Ben Abdelkhalek et al. Genes Dev. 2004.
Abstract
The floating head (flh) gene in zebrafish encodes a homeodomain protein, which is essential for notochord formation along the entire body axis. flh orthologs, termed Not genes, have been isolated from chick and Xenopus, but no mammalian ortholog has yet been identified. Truncate (tc) is an autosomal recessive mutation in mouse that specifically disrupts the development of the caudal notochord. Here, we demonstrate that truncate arose by a mutation in the mouse Not gene. The truncate allele (Nottc) contains a point mutation in the homeobox of Not that changes a conserved Phenylalanine residue in helix 1 to a Cysteine (F20C), and significantly destabilizes the homeodomain. Reversion of F20C in one allele of homozygous tc embryonic stem (ES) cells is sufficient to restore normal notochord formation in completely ES cell-derived embryos. We have generated a targeted mutation of Not by replacing most of the Not coding sequence, including the homeobox with the eGFP gene. The phenotype of NoteGFP/eGFP, NoteGFP/tc, and Nottc/tc embryos is very similar but slightly more severe in NoteGFP/eGFP than in Nottc/tc embryos. This confirms allelism of truncate and Not, and indicates that tc is not a complete null allele. Not expression is abolished in Foxa2 and T mutant embryos, suggesting that Not acts downstream of both genes during notochord development. This is in contrast to zebrafish embryos, in which flh interacts with ntl (zebrafish T) in a regulatory loop and is essential for development of the entire notochord, and suggests that different genetic control circuits act in different vertebrate species during notochord formation.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures
Figure 1.
Not localization, structure, and similarity to other vertebrate Not genes. (A) Physical map of the truncate region. The position and number of recombination breakpoints that were obtained in MOLF and CAST back-cross animals is indicated by ×s above the map, relevant BAC clones are shown below. (B) Structure of the Not gene. Boxes indicate exons. Black and white filling represents noncoding and coding regions, respectively. The homeobox is hatched. (C) Alignment of the homeodomains of mouse, chick, Xenopus, and zebrafish Not, and mouse Emx1 and Emx2 genes. The percentage of identical amino acids is indicated to the right. (D) Midpoint rooted phylogenetic tree of vertebrate Not genes based on ClustalW aligned homeodomains.
Figure 2.
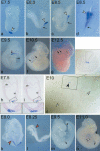
Expression of Not during embryonic development. Wild-type (a-g) and homozygous truncate (k-n) embryos and sections of wild-type (h-j) embryos after whole-mount in situ hybridization with a Not cDNA probe. Expression in wild-type embryos was first detected in the node, and was subsequently restricted to the node (arrowheads in a-d) and caudal portions of the notochord (arrows in b-g). (h,i) Sections of a hybridized day 7.5 embryo. The boxed regions showing the node in h and i are enlarged below. (j) Section of a day 10 embryo showing restriction of Not transcripts to the caudal notochord. No other expression domains were detected. White arrowheads in j point to the notochord in nonexpressing regions, the black arrowhead indicates the caudal _Not_-expressing notochord. In truncate embryos, ectopic transcripts were detected in the head process and anterior notochord (red arrowheads in k,l) of mutant day 8 and 8.25 embryos. Subsequently, the expression domain in the notochord (arrows in m,n) appeared extended. The arrowhead in n points to a gap in the notochord reflecting the tc phenotype. (ab) Allantoic bud; (hf) headfold.
Figure 3.
A point mutation in helix 1 of the homeodomain affects stability. (A) Partial nucleotide sequence of the wild-type and truncate Not allele around the T → G mutation. (B) Amino acid alignment of various homeodomains. An arrowhead indicates the position of the changed amino acid in Nottc. (C) Targeting strategy for reverting F20C. Exons are indicated by black boxes, relevant restriction sites and restriction fragments, as well as the probes used for genotyping, are shown above and below. The asterisk in exon 2 of the genomic locus indicates the point mutation. (D) Southern blot and PCR analysis of targeted clones after Cre-mediated excision of puro, using the primers ca1 and ca2 indicated in C. (E) Glycerol cleared wild-type (wt; panel a) and Nottc/tc (panel f) embryos collected from natural matings, and completely ES cell-derived embryos obtained with Nottc/tc (panels b,c,g,h) and Nottc/tcrev (panels d,e,i,j) cells, respectively, after in situ hybridization with a brachyury probe. Panels c, e, h, and j show higher magnifications of the embryos shown in panels b, d, g, and i. Arrowheads in panels c, f, and h point to gaps in the notochords. (F) UV-CD spectra obtained from HD NOT1-WT (solid line) and HD-NOT1-F20C (broken line). (G) Thermal denaturation curves obtained from HD NOT1-WT (open circles) and HD-NOT1-F20C (filled circles) monitored by the ellipticity of the absorption signal at 222 nm indicate a significant reduction of the melting temperature of HD-Not F20C (≈44°C compared with 57°C of the wild-type homeodomain).
Figure 4.
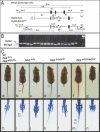
Gene targeting strategy and external and skeletal phenotypes of Not mutant mice. (A) Schematic representation of the genomic locus, targeting vector, and mutated allele. Exons are indicated by black boxes, relevant restriction sites and restriction fragments, as well as the probes used for genotyping are shown above and below. (B) Genotyping PCR on genomic DNA from newborns (two litters) derived from matings of NoteGFP/+ mice. (C) Representative examples of external adult phenotypes and skeletal preparations of Nottc/tc, NoteGFP/tc, and NoteGFP/eGFP newborn mice. Arrowheads point to constrictions in the tails and gaps in vertebrae, respectively.
Figure 5.
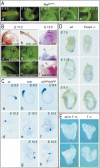
Notochord defects in Not mutant embryos and Not expression in embryos lacking Foxa2 or T function. (A) eGFP expression in heterozygous NoteGFP embryos between day 8.5 and 12.5 of development. (B) Notochord and tail defects in Nottc/tc (panels a-c), NoteGFP/tc (panels d-f), and NoteGFP/eGFP (panels g-i), day 11.5 (panels a,b,d,e,g,h) and 13.5 (panels c,f,i) embryos. Arrowheads in panels c, f, and i point to constrictions of the tails. In Nottc/tc day 11.5 embryos, the notochord was visualized by lacZ expression from a gene trap insertion into the Cobl locus, which was crossed into the mutant background. This insertion leads to lacZ expression in the notochord, but does not affect its development (Gasca et al. 1995). In Nottc/eGFP and NoteGFP/eGFP embryos, the notochord phenotype was assessed by eGFP fluorescence. All genotypes showed disruptions of the notochord in and caudal to the sacral region (arrows), or a discontinuous caudal notochord with scattered displaced notochord remnants (arrowheads). (C) Brachyury expression in day 9.5 (a-c) and 10.5 (d-h) wild-type (wt; panels a,d), Nottc/tc (panels b,e,g), and NoteGFP/eGFP (panels c,f,h) embryos. No notochord defects were observed in day 9.5 embryos. In day 10.5 embryos, notochords were apparently normal (panels e,f) or showed disruptions in the tail (panel g) or trunk region (panel h) indicated by arrowheads. (D,E) Absence of Not transcripts in _Foxa2_-/- (D) and _T_-/- (E) embryos.
Similar articles
- Spatial regulation of floating head expression in the developing notochord.
Melby AE, Kimelman D, Kimmel CB. Melby AE, et al. Dev Dyn. 1997 Jun;209(2):156-65. doi: 10.1002/(SICI)1097-0177(199706)209:2<156::AID-AJA2>3.0.CO;2-H. Dev Dyn. 1997. PMID: 9186051 - A homeobox gene essential for zebrafish notochord development.
Talbot WS, Trevarrow B, Halpern ME, Melby AE, Farr G, Postlethwait JH, Jowett T, Kimmel CB, Kimelman D. Talbot WS, et al. Nature. 1995 Nov 9;378(6553):150-7. doi: 10.1038/378150a0. Nature. 1995. PMID: 7477317 - Specification of cell fates at the dorsal margin of the zebrafish gastrula.
Melby AE, Warga RM, Kimmel CB. Melby AE, et al. Development. 1996 Jul;122(7):2225-37. doi: 10.1242/dev.122.7.2225. Development. 1996. PMID: 8681803 - Homeobox genes and orofacial development.
Sharpe PT. Sharpe PT. Connect Tissue Res. 1995;32(1-4):17-25. doi: 10.3109/03008209509013701. Connect Tissue Res. 1995. PMID: 7554914 Review. No abstract available. - The molecular and genetic analysis of mouse development.
Gossler A, Balling R. Gossler A, et al. Eur J Biochem. 1992 Feb 15;204(1):5-11. doi: 10.1111/j.1432-1033.1992.tb16599.x. Eur J Biochem. 1992. PMID: 1740155 Review.
Cited by
- Morphogenesis of the node and notochord: the cellular basis for the establishment and maintenance of left-right asymmetry in the mouse.
Lee JD, Anderson KV. Lee JD, et al. Dev Dyn. 2008 Dec;237(12):3464-76. doi: 10.1002/dvdy.21598. Dev Dyn. 2008. PMID: 18629866 Free PMC article. Review. - Nature and extent of left/right axis defects in T(Wis) /T(Wis) mutant mouse embryos.
Concepcion D, Papaioannou VE. Concepcion D, et al. Dev Dyn. 2014 Aug;243(8):1046-53. doi: 10.1002/dvdy.24144. Epub 2014 May 26. Dev Dyn. 2014. PMID: 24801048 Free PMC article. - Getting to the Core: Exploring the Embryonic Development from Notochord to Nucleus Pulposus.
Ambrosio L, Schol J, Ruiz-Fernández C, Tamagawa S, Joyce K, Nomura A, de Rinaldis E, Sakai D, Papalia R, Vadalà G, Denaro V. Ambrosio L, et al. J Dev Biol. 2024 Jul 3;12(3):18. doi: 10.3390/jdb12030018. J Dev Biol. 2024. PMID: 39051200 Free PMC article. Review. - In vitro modelling of anterior primitive streak patterning with human pluripotent stem cells identifies the path to notochord progenitors.
Robles-Garcia M, Thimonier C, Angoura K, Ozga E, MacPherson H, Blin G. Robles-Garcia M, et al. Development. 2024 Dec 15;151(24):dev202983. doi: 10.1242/dev.202983. Epub 2024 Dec 12. Development. 2024. PMID: 39611739 Free PMC article. - Clivus chordoma in continuity with a large pontine cyst.
Herold C, Giordano M, Naka T, Gerganov V, Samii M, Samii A. Herold C, et al. Skull Base. 2009 Mar;19(2):177-81. doi: 10.1055/s-0028-1096208. Skull Base. 2009. PMID: 19721775 Free PMC article.
References
- Ades S.E. and Sauer, R.T. 1994. Differential DNA-binding specificity of the engrailed homeodomain: The role of residue 50. Biochemistry 33: 9187-9194. - PubMed
- Amacher S.L. and Kimmel, C.B. 1998. Promoting notochord fate and repressing muscle development in zebrafish axial mesoderm. Development 125: 1397-1406. - PubMed
- Ang S.L. and Rossant, J. 1994. HNF-3b is essential for node and notochord formation in mouse development. Cell 78: 561-574. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials