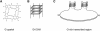Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA - PubMed (original) (raw)
Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA
Michelle L Duquette et al. Genes Dev. 2004.
Abstract
We show that intracellular transcription of G-rich regions produces novel DNA structures, visible by electron microscopy as large (150-500 bp) loops. These G-loops are formed cotranscriptionally, and they contain G4 DNA on one strand and a stable RNA/DNA hybrid on the other. G-loop formation requires a G-rich nontemplate strand and reflects the unusual stability of the rG/dC base pair. G-loops and G4 DNA form efficiently within plasmid genomes transcribed in vitro or in Escherichia coli. These results establish that G4 DNA can form in vivo, a finding with implications for stability and maintenance of all G-rich genomic regions.
Figures
Figure 1.
G4 DNA and G-quartets. (A) Four guanines assemble in a planar ring to form a G-quartet. (B) G4 DNA is a stable structure composed of stacked G-quartets (shaded squares). (C) Diagram of predicted structure formed upon transcription of G-rich DNA. The G-rich strand contains G4 DNA, stabilized by G-quartets; and the C-rich template strand is hybridized to the RNA transcript (gray). For simplicity, only two regions of G4 DNA structure are diagrammed, although loops formed upon transcription of a long G-rich region could, in principle, contain many structured regions.
Figure 2.
Loops form in transcribed G-rich DNAs. (A) Map of plasmid templates, which carry G-rich sequences downstream of a T7 promoter. (B) G-rich regions within the plasmid templates analyzed by EM. (C) Examples of loops formed upon transcription of pRX15F (Sμ repeat; left), pPH600 (Sγ3; middle), and pHumtel (telomeric repeat, right). DNA was transcribed in vitro, linearized with AflIII, and visualized by EM. Bar, 500 nm. (D) Maps of loops visualized in 15 randomly selected pRX15F (left), pPH600 (middle), and pHumtel (right) templates. Maps are drawn to scale.
Figure 3.
Loops contain an RNA/DNA hybrid formed cotranscriptionally. (A) Examples of loops formed in pHumtel transcribed with Dig-UTP and visualized with anti-Dig/gold beads. Bar, 200 nm. (B) Loops are destroyed by RNase H. Transcribed pRX15F before (left) and after (right) RNase H treatment. No loops were identified among 134 molecules visualized following treatment with RNase H. Bar, 500 nm. (C) Loops form cotranscriptionally. Treatment of pPH600 templates with 20 μg/mL RNase A during (example on left) or after (example on right) transcription did not alter loop formation; loops were evident in 58% (53/90) and 60% (60/99) of molecules, respectively. Bar, 500 nm.
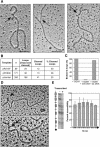
Figure 4.
Loops contain G4 DNA. (A) Examples of GQN1 cleavage of loops in transcribed pRX15F (Sμ; left), pPH600 (Sγ3; middle), and pHumtel (telomeric repeat, right). Arrows mark cleaved G-loops. Bar, 200 nm. (B) Effect of GQN1 treatment on transcribed pRX15F, pPH600, and pHumtel templates. DNAs were visualized by EM, and loops were scored as cleaved if they were opened and contained a clearly visible arm or ends, as in Figure 4A. (n) Number of molecules scored. (C) Effect of competitor G4 DNA (10-fold excess) or single-stranded DNA (1000-fold excess) on GQN1 cleavage of G-loops in transcribed plasmids, normalized to 100% cleavage in the absence of competitor. (D) G4 DNA recognized by recombinant biotinylated Nucleolin-428/streptavidin gold beads. Arrows indicate beads bound at loops. Bar, 200 nm. (E) DMS footprinting analysis verifies the presence of G-quartets. (Left) Example of a DMS footprint of pRG4F (left, mock transcribed; right, transcribed). (Right) compiled results of six independent footprinting experiments showing ratio of DMS accessibility of each G-run in the transcribed sample relative to the untranscribed control. Diagram indicates positions of G-runs (boxes) and single Gs (lines), and direction of transcription (arrows). Bars, S.D.
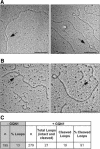
Figure 5.
Loops containing G4 DNA form in plasmids transcribed in E. coli. (A) Examples of loops formed in pPH600 (Sγ3) transcribed in E. coli strain NM 256 [AB1157 (λDE3) rnh::cat recQ::_kan_]. Bar, 200 nm. (B) Examples of GQN1 cleavage of loops formed in pPH600 (Sγ3) transcribed in E. coli NM 256. Arrows indicate broken loops. Bar, 200 nm. (C) Sensitivity of loops formed in E. coli to GQN1 cleavage. Loops were analyzed by EM and were scored as cleaved if they were opened and contained clearly visible DNA ends. (n) Number of molecules scored.
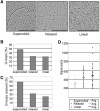
Figure 6.
Effect of topology on G-loop formation. (A) Examples of loops formed on supercoiled, relaxed, and linearized pPH600 templates. Bar, 500 nm. (B) Effect of plasmid topology on efficiency of loop formation. Loops were visualized by EM, and at least 400 molecules of each topoisomer were scored. (C) Effect of plasmid topology on G4 DNA formation. Templates of each topoisomer were treated with GQN1 and visualized by EM to assay cleavage; 200 GQN1-treated and 200-untreated molecules of each topoisomer were visualized. (D) Effect of plasmid topology on loop size. Loop size was measured following transcription of supercoiled, relaxed, or linear pPH600 templates, by EM visualization of 35 molecules of each topoisomer.
Similar articles
- G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter.
Roy D, Lieber MR. Roy D, et al. Mol Cell Biol. 2009 Jun;29(11):3124-33. doi: 10.1128/MCB.00139-09. Epub 2009 Mar 23. Mol Cell Biol. 2009. PMID: 19307304 Free PMC article. - AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation.
Duquette ML, Pham P, Goodman MF, Maizels N. Duquette ML, et al. Oncogene. 2005 Sep 1;24(38):5791-8. doi: 10.1038/sj.onc.1208746. Oncogene. 2005. PMID: 15940261 - Simultaneous probing of transcription, G-quadruplex, and R-loop.
Paul T, Yang L, Lee CY, Myong S. Paul T, et al. Methods Enzymol. 2024;705:377-396. doi: 10.1016/bs.mie.2024.07.004. Epub 2024 Aug 9. Methods Enzymol. 2024. PMID: 39389670 - R-loop structure: the formation and the effects on genomic stability.
Pan X, Jiang N, Chen X, Zhou X, Ding L, Duan F. Pan X, et al. Yi Chuan. 2014 Dec;36(12):1185-94. doi: 10.3724/SP.J.1005.2014.1185. Yi Chuan. 2014. PMID: 25487262 Review. - The Yin and Yang of R-loop biology.
Costantino L, Koshland D. Costantino L, et al. Curr Opin Cell Biol. 2015 Jun;34:39-45. doi: 10.1016/j.ceb.2015.04.008. Epub 2015 May 15. Curr Opin Cell Biol. 2015. PMID: 25938907 Free PMC article. Review.
Cited by
- Repeat instability during DNA repair: Insights from model systems.
Usdin K, House NC, Freudenreich CH. Usdin K, et al. Crit Rev Biochem Mol Biol. 2015 Mar-Apr;50(2):142-67. doi: 10.3109/10409238.2014.999192. Epub 2015 Jan 22. Crit Rev Biochem Mol Biol. 2015. PMID: 25608779 Free PMC article. Review. - G-quadruplex forming regions in GCK and TM6SF2 are targets for differential DNA methylation in metabolic disease and hepatocellular carcinoma patients.
Lahnsteiner A, Ellmer V, Oberlercher A, Liutkeviciute Z, Schönauer E, Paulweber B, Aigner E, Risch A. Lahnsteiner A, et al. Sci Rep. 2024 Aug 30;14(1):20215. doi: 10.1038/s41598-024-70749-0. Sci Rep. 2024. PMID: 39215018 Free PMC article. - G-Quadruplexes at Telomeres: Friend or Foe?
Bryan TM. Bryan TM. Molecules. 2020 Aug 13;25(16):3686. doi: 10.3390/molecules25163686. Molecules. 2020. PMID: 32823549 Free PMC article. Review. - Human telomere, oncogenic promoter and 5'-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics.
Patel DJ, Phan AT, Kuryavyi V. Patel DJ, et al. Nucleic Acids Res. 2007;35(22):7429-55. doi: 10.1093/nar/gkm711. Epub 2007 Oct 2. Nucleic Acids Res. 2007. PMID: 17913750 Free PMC article. Review. - Mechanism of R-loop formation at immunoglobulin class switch sequences.
Roy D, Yu K, Lieber MR. Roy D, et al. Mol Cell Biol. 2008 Jan;28(1):50-60. doi: 10.1128/MCB.01251-07. Epub 2007 Oct 22. Mol Cell Biol. 2008. PMID: 17954560 Free PMC article.
References
- Arakawa H., Iwasato, T., Hayashida, H., Shimizu, A., Honjo, T., and Yamagishi, H. 1993. The complete murine immunoglobulin class switch region of the α heavy chain gene-hierarchic repetitive structure and recombination breakpoints. J. Biol. Chem. 268: 4651-4655. - PubMed
- Bowater R.P. and Wells, R.D. 2001. The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. Mol. Biol. 66: 159-202. - PubMed
- Brown B.A., Lin, Y., Roberts, J.F., and Hardin, C.C. 1995. Antibodies specific for the DNA quadruplex [d(CGC G4 GCG)4] isolated from autoimmune mice. Nucleic Acids Symp Ser 33: 134-136. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM065988/GM/NIGMS NIH HHS/United States
- T32 HD007149/HD/NICHD NIH HHS/United States
- R01 GM39799/GM/NIGMS NIH HHS/United States
- GM31693/GM/NIGMS NIH HHS/United States
- R01 GM65988/GM/NIGMS NIH HHS/United States
- R01 GM031693/GM/NIGMS NIH HHS/United States
- T32 HD07149/HD/NICHD NIH HHS/United States
- R56 GM031693/GM/NIGMS NIH HHS/United States
- R01 GM039799/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources