A protein interaction framework for human mRNA degradation - PubMed (original) (raw)
Comparative Study
. 2004 Jul;14(7):1315-23.
doi: 10.1101/gr.2122004.
Affiliations
- PMID: 15231747
- PMCID: PMC442147
- DOI: 10.1101/gr.2122004
Comparative Study
A protein interaction framework for human mRNA degradation
Ben Lehner et al. Genome Res. 2004 Jul.
Abstract
The degradation of mRNA is an important regulatory step in the control of gene expression. However, mammalian RNA decay pathways remain poorly characterized. To provide a framework for studying mammalian RNA decay, a two-hybrid protein interaction map was generated using 54 constructs from 38 human proteins predicted to function in mRNA decay. The results provide evidence for interactions between many different proteins required for mRNA decay. Of particular interest are interactions between the poly(A) ribonuclease and the exosome and between the Lsm complex, decapping factors, and 5'-->3' exonucleases. Moreover, multiple interactions connect 5'-->3' and 3'-->5' decay proteins to each other and to nonsense-mediated decay factors, providing the opportunity for coordination between decay pathways. The interaction network also predicts the internal organization of the exosome and Lsm complexes. Additional interactions connect mRNA decay factors to many novel proteins and to proteins required for other steps in gene expression. These results provide an experimental insight into the organization of proteins required for mRNA decay and their coupling to other cellular processes, and the physiological relevance of many of these interactions are supported by their evolutionary conservation. The interactions also provide a wealth of hypotheses to guide future research on mRNA degradation and demonstrate the power of exhaustive protein interaction mapping in aiding understanding of uncharacterized protein complexes and pathways.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures
Figure 1
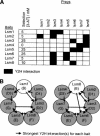
Subunit interactions of the human exosome. (A) Y2H interactions detected on stringent (–Ade) and less-stringent (–His) selection are indicated. The amino acid residues included in protein fragments are shown in brackets. (*) Self-active bait. (B) Stringent Y2H interactions are superimposed on a model of the exosome. The Rrp41-PM/Scl-75 interaction was only detected by library screening (see Table 2). Interactions previously detected by mammalian two-hybrid (M2H) and between the homologous yeast proteins are indicated. PH-domain subunits are blue, S1-domain subunits are red, and other subunits are yellow.
Figure 1
Subunit interactions of the human exosome. (A) Y2H interactions detected on stringent (–Ade) and less-stringent (–His) selection are indicated. The amino acid residues included in protein fragments are shown in brackets. (*) Self-active bait. (B) Stringent Y2H interactions are superimposed on a model of the exosome. The Rrp41-PM/Scl-75 interaction was only detected by library screening (see Table 2). Interactions previously detected by mammalian two-hybrid (M2H) and between the homologous yeast proteins are indicated. PH-domain subunits are blue, S1-domain subunits are red, and other subunits are yellow.
Figure 2
Subunit interactions of the human Lsm complexes. (A) Each potential Lsm–Lsm interaction was assayed on selective media lacking His and containing 0 mM, 5 mM, 10 mM, 25 mM, or 50 mM 3-AT, a competitive inhibitor of the HIS3 reporter. The interactions for each Lsm bait construct are shown at the indicated concentration of 3-AT, whereby no more than two interactions are seen with proteins from either the expected Lsm1–7 or Lsm2–8 complexes. (B) These interactions are shown on models of the Lsm1–7 and Lsm2–8 complexes. The most similar Sm protein is indicated in brackets for each Lsm protein.
Figure 3
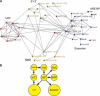
Protein–protein interactions detected between proteins predicted to function in human RNA decay. (A) Each Y2H interaction (selected on media lacking Ade or His) is indicated as an arrow from bait to prey. The amino acid residues included in protein fragments are indicated in brackets. Very weak interactions are not shown. The figure was drawn using BioLayout (
http://maine.ebi.ac.uk:8000/services/biolayout/
; Enright and Ouzounis 2001). (B) The interactions of the Upf proteins suggest how both nuclear and cytoplasmic 5′→3′ and 3′→5′ decay factors may be recruited during NMD. Y2H interactions are shown as arrows, proteins required for NMD are indicated by *, and proteins that coprecipitate from cell extracts are underlined (Lejeune et al. 2003).
Similar articles
- Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways.
van Dijk EL, Schilders G, Pruijn GJ. van Dijk EL, et al. RNA. 2007 Jul;13(7):1027-35. doi: 10.1261/rna.575107. Epub 2007 Jun 1. RNA. 2007. PMID: 17545563 Free PMC article. - Yeast Sm-like proteins function in mRNA decapping and decay.
Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Tharun S, et al. Nature. 2000 Mar 30;404(6777):515-8. doi: 10.1038/35006676. Nature. 2000. PMID: 10761922 - Zero tolerance for nonsense: nonsense-mediated mRNA decay uses multiple degradation pathways.
Wormington M. Wormington M. Mol Cell. 2003 Sep;12(3):536-8. doi: 10.1016/s1097-2765(03)00362-9. Mol Cell. 2003. PMID: 14527400 Review. No abstract available. - Cell and molecular biology of the exosome: how to make or break an RNA.
Schilders G, van Dijk E, Raijmakers R, Pruijn GJ. Schilders G, et al. Int Rev Cytol. 2006;251:159-208. doi: 10.1016/S0074-7696(06)51005-8. Int Rev Cytol. 2006. PMID: 16939780 Review.
Cited by
- Structure and Activities of the Eukaryotic RNA Exosome.
Wasmuth EV, Lima CD. Wasmuth EV, et al. Enzymes. 2012;31:53-75. doi: 10.1016/B978-0-12-404740-2.00003-3. Epub 2012 Sep 29. Enzymes. 2012. PMID: 27166440 Free PMC article. - Variants in EXOSC9 Disrupt the RNA Exosome and Result in Cerebellar Atrophy with Spinal Motor Neuronopathy.
Burns DT, Donkervoort S, Müller JS, Knierim E, Bharucha-Goebel D, Faqeih EA, Bell SK, AlFaifi AY, Monies D, Millan F, Retterer K, Dyack S, MacKay S, Morales-Gonzalez S, Giunta M, Munro B, Hudson G, Scavina M, Baker L, Massini TC, Lek M, Hu Y, Ezzo D, AlKuraya FS, Kang PB, Griffin H, Foley AR, Schuelke M, Horvath R, Bönnemann CG. Burns DT, et al. Am J Hum Genet. 2018 May 3;102(5):858-873. doi: 10.1016/j.ajhg.2018.03.011. Am J Hum Genet. 2018. PMID: 29727687 Free PMC article. - Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network.
Markson G, Kiel C, Hyde R, Brown S, Charalabous P, Bremm A, Semple J, Woodsmith J, Duley S, Salehi-Ashtiani K, Vidal M, Komander D, Serrano L, Lehner P, Sanderson CM. Markson G, et al. Genome Res. 2009 Oct;19(10):1905-11. doi: 10.1101/gr.093963.109. Epub 2009 Jun 23. Genome Res. 2009. PMID: 19549727 Free PMC article. - Exome sequencing in a family with intellectual disability, early onset spasticity, and cerebellar atrophy detects a novel mutation in EXOSC3.
Zanni G, Scotton C, Passarelli C, Fang M, Barresi S, Dallapiccola B, Wu B, Gualandi F, Ferlini A, Bertini E, Wei W. Zanni G, et al. Neurogenetics. 2013 Nov;14(3-4):247-50. doi: 10.1007/s10048-013-0371-z. Epub 2013 Aug 24. Neurogenetics. 2013. PMID: 23975261 - Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo.
D'Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, Baralle FE. D'Ambrogio A, et al. Nucleic Acids Res. 2009 Jul;37(12):4116-26. doi: 10.1093/nar/gkp342. Epub 2009 May 8. Nucleic Acids Res. 2009. PMID: 19429692 Free PMC article.
References
- Chen, C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., and Karin, M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464. - PubMed
- Collins, B.M., Cubeddu, L., Naidoo, N., Harrop, S.J., Kornfeld, G.D., Dawes, I.W., Curmi, P.M., and Mabbutt, B.C. 2003. Homomeric ring assemblies of eukaryotic Sm proteins have affinity for both RNA and DNA. Crystal structure of an oligomeric complex of yeast SmF. J. Biol. Chem. 278: 17291–17298. - PubMed
WEB SITE REFERENCES
- http://maine.ebi.ac.uk:8000/services/biolayout; Biolayout software.
- http://www.hgmp.mrc.ac.uk/Research/RNA; descriptions of all interacting proteins isolated in this study.
- http://inparanoid.cgb.ki.se/; The Inparanoid database of pairwise orthologs.
- http://www.blueprint.org/bind/bind.php; BIND protein interaction database.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases