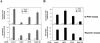Functional proteomics mapping of a human signaling pathway - PubMed (original) (raw)
Functional proteomics mapping of a human signaling pathway
Frédéric Colland et al. Genome Res. 2004 Jul.
Abstract
Access to the human genome facilitates extensive functional proteomics studies. Here, we present an integrated approach combining large-scale protein interaction mapping, exploration of the interaction network, and cellular functional assays performed on newly identified proteins involved in a human signaling pathway. As a proof of principle, we studied the Smad signaling system, which is regulated by members of the transforming growth factor beta (TGFbeta) superfamily. We used two-hybrid screening to map Smad signaling protein-protein interactions and to establish a network of 755 interactions, involving 591 proteins, 179 of which were poorly or not annotated. The exploration of such complex interaction databases is improved by the use of PIMRider, a dedicated navigation tool accessible through the Web. The biological meaning of this network is illustrated by the presence of 18 known Smad-associated proteins. Functional assays performed in mammalian cells including siRNA knock-down experiments identified eight novel proteins involved in Smad signaling, thus validating this integrated functional proteomics approach.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures
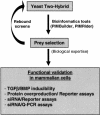
Figure 1
Strategy for unraveling interactions between components of the Smad pathway. This figure describes the global process from our large-scale yeast two-hybrid screening to the general functional validation assays in mammalian cells. Preys were selected by use of bioinformatics tools (PIMBuilder, PIMRider) and biological expertise.
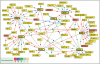
Figure 2
Protein interaction map around some selected baits in the TGFβ pathway. PBS D and E interactions as well as protein interactions corresponding to the BMP pathway have been excluded from this map for clarity (however, all data are available on the PIMRider visualization platform at
). The following bait proteins were selected to generate a TGFβ network: Smad2, 3, 4, 7, SARA, Smurf2, SnoN, and SNIP1. These are represented in boxes with heavy outlining. Of the 14 proteins selected for further functional validation, the six proteins present in this figure are represented in red. The (+) symbol located in the upper right corner of some boxes indicates that additional partners are not represented in this figure but are visible on the Web site mentioned above.
Figure 3
Involvement of LMO4, HYPA, and KIAA1196 in the Smad pathway. (A) Following siRNA-mediated cellular knock-down targeting LMO4, HYPA, and KIAA1196, Q-PCR and reporter assays were used to analyze endogenous levels of human alkaline phosphatase (AP) mRNA and BMP-induced reporter (BMP-RE) activity in HepG2 cells, respectively. AP mRNA levels and BMP-RE activity were determined in untreated (white) and BMP7-treated (gray) HepG2 cells. (B) Following siRNA-mediated cellular knock-down targeting LMO4, HYPA, and KIAA1196, Q-PCR and reporter assays were used to analyze endogenous levels of PAI-1 mRNA and TGFβ-induced reporter (TGFβ-RE) activity in HepG2 cells, respectively. PAI-1 mRNA levels and TGFβ-RE activity were determined both in untreated (white) and TGFβ-treated (black) HepG2 cells. In all Q-PCR experiments, mRNA levels were normalized according to an internal GUS control. The specific luciferase activity was normalized using the pRL-TK vector. All results are mean values ±SE calculated from triplicates performed in at least two independent experiments.
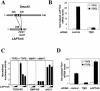
Figure 4
Involvement of LAPTm5 in the TGFβ pathway. (A) Interaction between Smurf2 and LAPTm5. The full-length proteins are represented in gray; black boxes correspond to the interaction domains. Positions are indicated in amino acids. White boxes correspond to WW motifs in Smurf2. The PPXY motif of LAPTm5 is indicated. (B) Endogenous levels of LAPTm5 mRNA were determined in HepG2 cells by Q-PCR in the absence (white) and the presence (black) of TGFβ for 18 h with or without a TβRI–targeting siRNA duplex. (C) The effect of LAPTm5 overproduction was studied using the following luciferase reporter vectors: a TGFβ-responsive element (TGFβ-RE), a BMP-responsive element (BMP-RE), and an unrelated reporter (pGL3 control). The effect was measured in the presence and the absence of TGFβ or BMP7. HepG2 cells were transfected with 0, 2, or 10 ng of pV3-LAPTm5. The specific luciferase activity was normalized using the pRL-TK vector. (D) Following siRNA-mediated cellular knock-down targeting TβRI and LAPTm5, Q-PCR was used to analyze the endogenous levels of PAI-1 mRNA both in untreated (white) and TGFβ-treated (black) HepG2 cells. mRNA levels were normalized according to an internal GUS control. All results are mean values ±SE calculated from triplicates performed in at least two independent experiments.
Similar articles
- Modulation of the interleukin-6 receptor subunit glycoprotein 130 complex and its signaling by LMO4 interaction.
Novotny-Diermayr V, Lin B, Gu L, Cao X. Novotny-Diermayr V, et al. J Biol Chem. 2005 Apr 1;280(13):12747-57. doi: 10.1074/jbc.M500175200. Epub 2005 Jan 27. J Biol Chem. 2005. PMID: 15677447 - Regulatory mechanisms for transforming growth factor beta as an autocrine inhibitor in human hepatocellular carcinoma: implications for roles of smads in its growth.
Matsuzaki K, Date M, Furukawa F, Tahashi Y, Matsushita M, Sugano Y, Yamashiki N, Nakagawa T, Seki T, Nishizawa M, Fujisawa J, Inoue K. Matsuzaki K, et al. Hepatology. 2000 Aug;32(2):218-27. doi: 10.1053/jhep.2000.9145. Hepatology. 2000. PMID: 10915727 - SIP1 (Smad interacting protein 1) and deltaEF1 (delta-crystallin enhancer binding factor) are structurally similar transcriptional repressors.
van Grunsven LA, Schellens A, Huylebroeck D, Verschueren K. van Grunsven LA, et al. J Bone Joint Surg Am. 2001;83-A Suppl 1(Pt 1):S40-7. J Bone Joint Surg Am. 2001. PMID: 11263664 Review. - Functional interaction between Smad, CREB binding protein, and p68 RNA helicase.
Warner DR, Bhattacherjee V, Yin X, Singh S, Mukhopadhyay P, Pisano MM, Greene RM. Warner DR, et al. Biochem Biophys Res Commun. 2004 Nov 5;324(1):70-6. doi: 10.1016/j.bbrc.2004.09.017. Biochem Biophys Res Commun. 2004. PMID: 15464984 - TGF-beta signaling by Smad proteins.
Miyazono K, ten Dijke P, Heldin CH. Miyazono K, et al. Adv Immunol. 2000;75:115-57. doi: 10.1016/s0065-2776(00)75003-6. Adv Immunol. 2000. PMID: 10879283 Review.
Cited by
- An aPKC-exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation.
Rosse C, Formstecher E, Boeckeler K, Zhao Y, Kremerskothen J, White MD, Camonis JH, Parker PJ. Rosse C, et al. PLoS Biol. 2009 Nov;7(11):e1000235. doi: 10.1371/journal.pbio.1000235. Epub 2009 Nov 3. PLoS Biol. 2009. PMID: 19885391 Free PMC article. - Analysis of single nucleotide polymorphisms in the region of CLDN2-MORC4 in relation to inflammatory bowel disease.
Söderman J, Norén E, Christiansson M, Bragde H, Thiébaut R, Hugot JP, Tysk C, O'Morain CA, Gassull M, Finkel Y, Colombel JF, Lémann M, Almer S. Söderman J, et al. World J Gastroenterol. 2013 Aug 14;19(30):4935-43. doi: 10.3748/wjg.v19.i30.4935. World J Gastroenterol. 2013. PMID: 23946598 Free PMC article. - Cell-type specific profiling of human entorhinal cortex at the onset of Alzheimer's disease neuropathology.
Rodriguez-Rodriguez P, Wang W, Tsagkogianni C, Feng I, Morello-Megias A, Jain K, Alanko V, Kahvecioglu HA, Mohammadi E, Li X, Flajolet M, Sandebring-Matton A, Maioli S, Vidal N, Milosevic A, Roussarie JP. Rodriguez-Rodriguez P, et al. bioRxiv [Preprint]. 2025 Feb 3:2024.12.31.630881. doi: 10.1101/2024.12.31.630881. bioRxiv. 2025. PMID: 39803521 Free PMC article. Preprint. - Risk of Alzheimer's disease in Down syndrome: Insights gained by multi-omics.
Qu HQ, Liu Y, Connolly JJ, Mentch FD, Kao C, Hakonarson H. Qu HQ, et al. Alzheimers Dement. 2025 Apr;21(4):e14604. doi: 10.1002/alz.14604. Alzheimers Dement. 2025. PMID: 40207399 Free PMC article. Review. - RFX1: a promising therapeutic arsenal against cancer.
Issac J, Raveendran PS, Das AV. Issac J, et al. Cancer Cell Int. 2021 May 8;21(1):253. doi: 10.1186/s12935-021-01952-6. Cancer Cell Int. 2021. PMID: 33964962 Free PMC article. Review.
References
- Adra, C.N., Zhu, S., Ko, J.L., Guillemot, J.C., Cuervo, A.M., Kobayashi, H., Horiuchi, T., Lelias, J.M., Rowley, J.D., and Lim, B. 1996. LAPTM5: A novel lysosomal-associated multispanning membrane protein preferentially expressed in hematopoietic cells. Genomics 35: 328–337. - PubMed
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D., et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37–40. - PMC - PubMed
- Attisano, L. and Wrana, J.L. 2002. Signal transduction by the TGF-β superfamily. Science 296: 1646–1647. - PubMed
WEB SITE REFERENCES
- http://pim.hybrigenics.com; PIMRider.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials