Global survey of chromatin accessibility using DNA microarrays - PubMed (original) (raw)
Global survey of chromatin accessibility using DNA microarrays
M Ryan Weil et al. Genome Res. 2004 Jul.
Abstract
An increasing number of studies indicate a central role for chromatin remodeling in the regulation of gene expression. Current methods for high-resolution studies of the relationship between chromatin accessibility and transcription are low throughput, making a genome-wide study impractical. To enable the simultaneous measurement of the global chromatin accessibility state at the resolution of single genes, we developed the Chromatin Array technique, in which chromatin is separated by its condensation state using either the solubility differences of mono- and oligonucleosomes in specific buffers or controlled DNase I digestion and selection of the large refractory (condensed) DNA fragments. By probing with a comparative genomic hybridization style microarray, we can determine the condensation state of thousands of individual loci and correlate this with transcriptional activity. Applying this technique to the breast tumor model cell line, MCF7, we found that when the condensation is homogeneous in the population of cells, expression is inversely proportional to the level of accessibility and the two methods of accessibility-based target selection correlate well. Using functional annotation and comparative genomic hybridization data, we have begun to decipher the possible biological implications of the relationship between chromatin accessibility and expression.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures
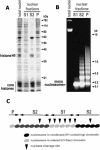
Figure 1
The chromatin solubility assay is used to extract the S1, S2, and P fractions, and the result of that process is depicted as the patterns seen in the gels of the resulting protein and DNA. The protein gel (A) shows the depletion of the H1 histone and the enrichment of nonhistone proteins in the S1 and P fractions. The DNA gel (B) shows the difference in the extent of the digestion between the fractions. The model (C) illustrates the basic concept of the chromatin fractionation assay.
Figure 2
The log scale scatter plot of the co-normalized intensities (sequence abundances) illustrates the relationship between condensation (the ratio of the S2 fraction intensity to total genomic DNA intensity) and the absolute RNA expression. The light gray triangles are Group 2, which has a condensation to expression (C/E) ratio of <0.5. The black squares represent Group 3, which has indeterminate accessibility and a C/E ratio of <2.0 to >0.5. The gray diamonds represent Group 1, which has a C/E ratio of >2.0.
Figure 3
A fragment length selection gel showing the distribution of DNase I-cleaved fragments is used to identify the upper 40% of the total lane intensity to direct the excision of the packing enriched region. The lane digested with the highest concentration of DNase I (64 U) was chosen for its uniform distribution of fragments around the 5-kb median. This section of the gel was excised, and the DNA was recovered for labeling. (M) λ HindIII DNA mass standard.
Similar articles
- Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells.
Lin CY, Ström A, Vega VB, Kong SL, Yeo AL, Thomsen JS, Chan WC, Doray B, Bangarusamy DK, Ramasamy A, Vergara LA, Tang S, Chong A, Bajic VB, Miller LD, Gustafsson JA, Liu ET. Lin CY, et al. Genome Biol. 2004;5(9):R66. doi: 10.1186/gb-2004-5-9-r66. Epub 2004 Aug 12. Genome Biol. 2004. PMID: 15345050 Free PMC article. - Improving signal intensities for genes with low-expression on oligonucleotide microarrays.
Ramdas L, Cogdell DE, Jia JY, Taylor EE, Dunmire VR, Hu L, Hamilton SR, Zhang W. Ramdas L, et al. BMC Genomics. 2004 Jun 14;5(1):35. doi: 10.1186/1471-2164-5-35. BMC Genomics. 2004. PMID: 15196312 Free PMC article. - Isolation of nuclei for use in genome-wide DNase hypersensitivity assays to probe chromatin structure.
Ling G, Waxman DJ. Ling G, et al. Methods Mol Biol. 2013;977:13-9. doi: 10.1007/978-1-62703-284-1_2. Methods Mol Biol. 2013. PMID: 23436350 Free PMC article. - From correlation to causality: microarrays, cancer, and cancer treatment.
O'Neill GM, Catchpoole DR, Golemis EA. O'Neill GM, et al. Biotechniques. 2003 Mar;Suppl:64-71. Biotechniques. 2003. PMID: 12664688 Review. No abstract available. - Genomic profiling: cDNA arrays and oligoarrays.
Gorreta F, Carbone W, Barzaghi D. Gorreta F, et al. Methods Mol Biol. 2012;823:89-105. doi: 10.1007/978-1-60327-216-2_7. Methods Mol Biol. 2012. PMID: 22081341 Review.
Cited by
- Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin.
Johnson SM, Tan FJ, McCullough HL, Riordan DP, Fire AZ. Johnson SM, et al. Genome Res. 2006 Dec;16(12):1505-16. doi: 10.1101/gr.5560806. Epub 2006 Oct 12. Genome Res. 2006. PMID: 17038564 Free PMC article. - Applying whole-genome studies of epigenetic regulation to study human disease.
Lieb JD, Beck S, Bulyk ML, Farnham P, Hattori N, Henikoff S, Liu XS, Okumura K, Shiota K, Ushijima T, Greally JM. Lieb JD, et al. Cytogenet Genome Res. 2006;114(1):1-15. doi: 10.1159/000091922. Cytogenet Genome Res. 2006. PMID: 16717444 Free PMC article. Review. No abstract available. - Patterns in the genome.
Bickmore WA. Bickmore WA. Heredity (Edinb). 2019 Jul;123(1):50-57. doi: 10.1038/s41437-019-0220-4. Epub 2019 Jun 12. Heredity (Edinb). 2019. PMID: 31189906 Free PMC article. Review. - Reversing chromatin accessibility differences that distinguish homologous mitotic metaphase chromosomes.
Khan WA, Rogan PK, Knoll JH. Khan WA, et al. Mol Cytogenet. 2015 Aug 13;8:65. doi: 10.1186/s13039-015-0159-y. eCollection 2015. Mol Cytogenet. 2015. PMID: 26273322 Free PMC article. - Distinct modes of DNA accessibility in plant chromatin.
Shu H, Wildhaber T, Siretskiy A, Gruissem W, Hennig L. Shu H, et al. Nat Commun. 2012;3:1281. doi: 10.1038/ncomms2259. Nat Commun. 2012. PMID: 23232411
References
- Albertson, D.G., Collins, C., McCormick, F., and Gray, J.W. 2003. Chromosome aberrations in solid tumors. Nat. Genet. 34: 369–376. - PubMed
- Banerjee, S. and Hulten, M.A. 1994. Sperm nuclear chromatin transformations in somatic cell-free extracts. Mol. Reprod. Dev. 37: 305–317. - PubMed
- Banerjee, D., Mayer-Kuckuk, P., Capiaux, G., Budak-Alpdogan, T., Gorlick, R., and Bertino, J.R. 2002. Novel aspects of resistance to drugs targeted to dihydrofolate reductase and thymidylate synthase. Biochim. Biophys. Acta 1587: 164–173. - PubMed
WEB SITE REFERENCES
- http://famine.swmed.edu; the Garner lab GeneTraffic Server.
Publication types
MeSH terms
Substances
Grants and funding
- R33-CA81656/CA/NCI NIH HHS/United States
- T32-HL07360/HL/NHLBI NIH HHS/United States
- T32 HL007360/HL/NHLBI NIH HHS/United States
- 50CA70907/CA/NCI NIH HHS/United States
- P50 CA070907/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources