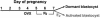Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation - PubMed (original) (raw)
Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation
Toshio Hamatani et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Delayed implantation (embryonic diapause) occurs when the embryo at the blastocyst stage achieves a state of suspended animation. During this period, blastocyst growth is very slow, with minimal or no cell division. Nearly 100 mammals in seven different orders undergo delayed implantation, but the underlying molecular mechanisms that direct this process remain largely unknown. In mice, ovariectomy before preimplantation ovarian estrogen secretion on day 4 of pregnancy initiates blastocyst dormancy, which normally lasts for 1-2 weeks by continued progesterone treatment, although blastocyst survival decreases with time. An estrogen injection rapidly activates blastocysts and initiates their implantation in the progesterone-primed uterus. Using this model, here we show that among approximately 20,000 genes examined, only 229 are differentially expressed between dormant and activated blastocysts. The major functional categories of altered genes include the cell cycle, cell signaling, and energy metabolic pathways, particularly highlighting the importance of heparin-binding epidermal growth factor-like signaling in blastocyst-uterine crosstalk in implantation. The results provide evidence that the two different physiological states of the blastocyst, dormancy and activation, are molecularly distinguishable in a global perspective and underscore the importance of specific molecular pathways in these processes. This study has identified candidate genes that provide a scope for in-depth analysis of their functions and an opportunity for examining their relevance to blastocyst dormancy and activation in numerous other species for which microarray analysis is not available or possible due to very limited availability of blastocysts.
Figures
Fig. 1.
Experimental design to generate dormant and activated blastocysts. Pregnant mice were ovariectomized on the morning (0900 h) of day 4. Delayed implantation was maintained by daily injection of P4 from days 5–7. To activate dormant blastocysts, P4-primed implanting mice were injected with estradiol-17β(_E_2) on day 7. Blastocysts were recovered between 12 and 14 h after the last steroid injections. OVX, ovariectomy.
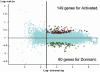
Fig. 2.
A scatter plot of 21,939 gene features on NIA 22,000 60-mer oligo microarray, comparing gene expression in dormant and activated blastocysts. Microarray data were analyzed by ANOVA-FDR statistics. The combined results of six hybridizations identified 229 differentially expressed genes (averaged log intensity ≥2.3, P ≤ 0.01, and FDR ≤0.1), including 80 genes highly expressed in dormant blastocysts (green circles) and 149 genes highly expressed in activated blastocysts (red circles).
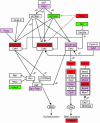
Fig. 3.
A chart of cell cycle genes. Green and red boxes highlight for higher expression (FDR < 0.1) in dormant and activated blastocysts, respectively. The genes, which showed higher expression with P ≤ 0.05 and FDR ≥ 0.1 in activated blastocysts, are highlighted as pink boxes. The lines with arrowheads or T heads represent stimulation or suppression, respectively. The pathway figure is based on the Kyoto Encyclopedia of Genes and Genomes pathway database (49), with slight modification. Based on published data (16), the dashed lines from Brca1 represent increased expression of p21cip1/WAF1 and decreased expression of cyclin E in _Brca1_–/– embryos. +P, –P, and +U indicate phosphorylation, dephosphorylation, and ubiquitination, respectively.
Fig. 4.
Localization of BRCA1, HB-EGF, p21, and IGF2R in dormant and activated blastocysts. Antigens (green) and nuclei (red) were visualized by using FITC-conjugated secondary antibodies and propidium iodide, respectively. Note higher signal intensity for BRCA1 and HB-EGF in activated blastocysts and of p21 and IGF2R in dormant blastocysts. (Bar = 50 μm.)
Fig. 5.
In situ hybridization of Hegf1 at the implantation or bead sites. (A) A representative section of a day 5 implantation site from natural mating. (B and C) A representative uterine section containing beads preabsorbed in BSA (B) or in HB-EGF (C). Arrow indicates the location of a blastocyst, whereas arrowheads indicate the location of beads (×100).
Similar articles
- Blastocyst's state of activity determines the "window" of implantation in the receptive mouse uterus.
Paria BC, Huet-Hudson YM, Dey SK. Paria BC, et al. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10159-62. doi: 10.1073/pnas.90.21.10159. Proc Natl Acad Sci U S A. 1993. PMID: 8234270 Free PMC article. - Coordination of differential effects of primary estrogen and catecholestrogen on two distinct targets mediates embryo implantation in the mouse.
Paria BC, Lim H, Wang XN, Liehr J, Das SK, Dey SK. Paria BC, et al. Endocrinology. 1998 Dec;139(12):5235-46. doi: 10.1210/endo.139.12.6386. Endocrinology. 1998. PMID: 9832464 - Intracellular signaling in the developing blastocyst as a consequence of the maternal-embryonic dialogue.
Armant DR, Wang J, Liu Z. Armant DR, et al. Semin Reprod Med. 2000;18(3):273-87. doi: 10.1055/s-2000-12565. Semin Reprod Med. 2000. PMID: 11299966 Review. - Molecular interactions at the maternal-embryonic interface during the early phase of implantation.
Kimber SJ. Kimber SJ. Semin Reprod Med. 2000;18(3):237-53. doi: 10.1055/s-2000-12562. Semin Reprod Med. 2000. PMID: 11299963 Review.
Cited by
- Metabolic Control over mTOR-Dependent Diapause-like State.
Hussein AM, Wang Y, Mathieu J, Margaretha L, Song C, Jones DC, Cavanaugh C, Miklas JW, Mahen E, Showalter MR, Ruzzo WL, Fiehn O, Ware CB, Blau CA, Ruohola-Baker H. Hussein AM, et al. Dev Cell. 2020 Jan 27;52(2):236-250.e7. doi: 10.1016/j.devcel.2019.12.018. Dev Cell. 2020. PMID: 31991105 Free PMC article. - Lipid droplets in mammalian eggs are utilized during embryonic diapause.
Arena R, Bisogno S, Gąsior Ł, Rudnicka J, Bernhardt L, Haaf T, Zacchini F, Bochenek M, Fic K, Bik E, Barańska M, Bodzoń-Kułakowska A, Suder P, Depciuch J, Gurgul A, Polański Z, Ptak GE. Arena R, et al. Proc Natl Acad Sci U S A. 2021 Mar 9;118(10):e2018362118. doi: 10.1073/pnas.2018362118. Proc Natl Acad Sci U S A. 2021. PMID: 33649221 Free PMC article. - Molecular mechanisms involved in progesterone receptor regulation of uterine function.
Lee K, Jeong J, Tsai MJ, Tsai S, Lydon JP, DeMayo FJ. Lee K, et al. J Steroid Biochem Mol Biol. 2006 Dec;102(1-5):41-50. doi: 10.1016/j.jsbmb.2006.09.006. Epub 2006 Oct 25. J Steroid Biochem Mol Biol. 2006. PMID: 17067792 Free PMC article. Review. - Embryo-derive TNF promotes decidualization via fibroblast activation.
Chen ST, Shi WW, Lin YQ, Yang ZS, Wang Y, Li MY, Li Y, Liu AX, Hu Y, Yang ZM. Chen ST, et al. Elife. 2023 Jul 17;12:e82970. doi: 10.7554/eLife.82970. Elife. 2023. PMID: 37458359 Free PMC article. - Restraint stress inhibits mouse implantation: temporal window and the involvement of HB-EGF, estrogen and progesterone.
Zhao LH, Cui XZ, Yuan HJ, Liang B, Zheng LL, Liu YX, Luo MJ, Tan JH. Zhao LH, et al. PLoS One. 2013 Nov 14;8(11):e80472. doi: 10.1371/journal.pone.0080472. eCollection 2013. PLoS One. 2013. PMID: 24244689 Free PMC article.
References
- Dey, S. K., Lim, H., Das, S. K., Reese, J., Paria, B. C., Daikoku, T. & Wang, H. (2004) Endocr. Rev. 25, 341–373. - PubMed
- Paria, B. C., Reese, J., Das, S. K. & Dey, S. K. (2002) Science 296, 2185–2188. - PubMed
- Yoshinaga, K. & Adams, C. E. (1966) J. Reprod. Fertil. 12, 593–595. - PubMed
- McLaren, A. (1968) J. Endocrinol. 42, 453–463. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DA 06668/DA/NIDA NIH HHS/United States
- P30 HD033994/HD/NICHD NIH HHS/United States
- R37 HD012304/HD/NICHD NIH HHS/United States
- HD12304/HD/NICHD NIH HHS/United States
- R01 DA006668/DA/NIDA NIH HHS/United States
- R37 DA006668/DA/NIDA NIH HHS/United States
- U54 HD033994/HD/NICHD NIH HHS/United States
- HD33994/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases