A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination - PubMed (original) (raw)
A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination
Stella A Martomo et al. J Exp Med. 2004.
Abstract
Somatic hypermutation is initiated by activation-induced cytidine deaminase (AID), and occurs in several kilobases of DNA around rearranged immunoglobulin variable (V) genes and switch (S) sites before constant genes. AID deaminates cytosine to uracil, which can produce mutations of C:G nucleotide pairs, and the mismatch repair protein Msh2 participates in generating substitutions of downstream A:T pairs. Msh2 is always found as a heterodimer with either Msh3 or Msh6, so it is important to know which one is involved. Therefore, we sequenced V and S regions from Msh3- and Msh6-deficient mice and compared mutations to those from wild-type mice. Msh6-deficient mice had fewer substitutions of A and T bases in both regions and reduced heavy chain class switching, whereas Msh3-deficient mice had normal antibody responses. This establishes a role for the Msh2-Msh6 heterodimer in hypermutation and switch recombination. When the positions of mutation were mapped, several focused peaks were found in Msh6(-/-) clones, whereas mutations were dispersed in Msh3(-/-) and wild-type clones. The peaks occurred at either G or C in WGCW motifs (W = A or T), indicating that C was mutated on both DNA strands. This suggests that AID has limited entry points into V and S regions in vivo, and subsequent mutation requires Msh2-Msh6 and DNA polymerase.
Figures
Figure 1.
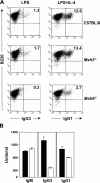
Diminished heavy chain class switching in Msh6 −/− but not Msh3 −/− B cells. (A) Flow cytometry analysis. Spleen cells were stimulated with LPS or LPS plus IL-4 to induce switching, and surface immunoglobulin was measured 3 d later. The percentage of B220+ cells that switched to IgG3 or IgG1 is shown in each box. (B) Serum antibody analysis. An ELISA was used to quantify anti-KLH antibodies from immunized mice. Black bars, C57BL/6; white bars, Msh6 −/−. Units are defined as the antibody titer that was normalized to the total amount of each isotype.
Figure 2.
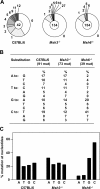
Fewer A:T substitutions in JH4 introns from Msh6 −/− clones. (A) The total number of clones analyzed is shown in the center of each circle. Segments represent the proportion of clones that contain the indicated number of mutations. (B) The sequence contains 26% A, 32% T, 28% G, and 14% C; values were corrected to represent a sequence with equal amounts of the four nucleotides. Mutations are shown from the nontranscribed strand. (C) Total mutations for each nucleotide are grouped.
Figure 3.
Reduced A:T mutations in Sμ from Msh6 −/− clones. (A) The proportion of clones containing mutations is shown. (B) Values are corrected for nucleotide composition of the sequence, which contained 33% A, 23% T, 28% G, and 16% C. (C) Total mutations of each nucleotide are grouped.
Figure 4.
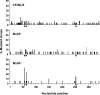
Distribution of mutations in JH4 intron reveals peaks in Msh6 −/− clones. Nucleotides are numbered from the first base after the JH4 coding sequence, with 1 corresponding to 2,340 of Genbank/EMBL/DDBJ under accession no. J00440. Data were calculated as the number of mutations at a nucleotide position divided by the number of mutated clones. Bars below the abscissa depict WGCW motifs.
Figure 5.
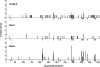
Location of mutations in Sμ shows targeting in Msh6 −/− clones. Nucleotides are numbered with 1 corresponding to 4,600 of Genbank/EMBL/DDBJ under accession no. J00440. Data represent the number of mutations at a position divided by the total number of clones. Bars below the abscissa represent WGCW motifs.
Similar articles
- The mismatch repair protein Msh6 influences the in vivo AID targeting to the Ig locus.
Li Z, Zhao C, Iglesias-Ussel MD, Polonskaya Z, Zhuang M, Yang G, Luo Z, Edelmann W, Scharff MD. Li Z, et al. Immunity. 2006 Apr;24(4):393-403. doi: 10.1016/j.immuni.2006.02.011. Immunity. 2006. PMID: 16618598 - Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation.
Ehrenstein MR, Neuberger MS. Ehrenstein MR, et al. EMBO J. 1999 Jun 15;18(12):3484-90. doi: 10.1093/emboj/18.12.3484. EMBO J. 1999. PMID: 10369687 Free PMC article. - Examination of Msh6- and Msh3-deficient mice in class switching reveals overlapping and distinct roles of MutS homologues in antibody diversification.
Li Z, Scherer SJ, Ronai D, Iglesias-Ussel MD, Peled JU, Bardwell PD, Zhuang M, Lee K, Martin A, Edelmann W, Scharff MD. Li Z, et al. J Exp Med. 2004 Jul 5;200(1):47-59. doi: 10.1084/jem.20040355. J Exp Med. 2004. PMID: 15238604 Free PMC article. - Activation-induced cytidine deaminase: a dual role in class-switch recombination and somatic hypermutation.
Durandy A. Durandy A. Eur J Immunol. 2003 Aug;33(8):2069-73. doi: 10.1002/eji.200324133. Eur J Immunol. 2003. PMID: 12884279 Review. - An update on the role of translesion synthesis DNA polymerases in Ig hypermutation.
Diaz M, Lawrence C. Diaz M, et al. Trends Immunol. 2005 Apr;26(4):215-20. doi: 10.1016/j.it.2005.02.008. Trends Immunol. 2005. PMID: 15797512 Review.
Cited by
- Identifying protein-protein interactions in somatic hypermutation.
Goodman MF, Scharff MD. Goodman MF, et al. J Exp Med. 2005 Feb 21;201(4):493-6. doi: 10.1084/jem.20050161. Epub 2005 Feb 14. J Exp Med. 2005. PMID: 15710655 Free PMC article. - Controlling somatic hypermutation in immunoglobulin variable and switch regions.
Maul RW, Gearhart PJ. Maul RW, et al. Immunol Res. 2010 Jul;47(1-3):113-22. doi: 10.1007/s12026-009-8142-5. Immunol Res. 2010. PMID: 20082153 Free PMC article. Review. - MSH2/MSH6 complex promotes error-free repair of AID-induced dU:G mispairs as well as error-prone hypermutation of A:T sites.
Roa S, Li Z, Peled JU, Zhao C, Edelmann W, Scharff MD. Roa S, et al. PLoS One. 2010 Jun 17;5(6):e11182. doi: 10.1371/journal.pone.0011182. PLoS One. 2010. PMID: 20567595 Free PMC article. - No Overt Clinical Immunodeficiency Despite Immune Biological Abnormalities in Patients With Constitutional Mismatch Repair Deficiency.
Tesch VK, IJspeert H, Raicht A, Rueda D, Dominguez-Pinilla N, Allende LM, Colas C, Rosenbaum T, Ilencikova D, Baris HN, Nathrath MHM, Suerink M, Januszkiewicz-Lewandowska D, Ragab I, Azizi AA, Wenzel SS, Zschocke J, Schwinger W, Kloor M, Blattmann C, Brugieres L, van der Burg M, Wimmer K, Seidel MG. Tesch VK, et al. Front Immunol. 2018 Jul 2;9:1506. doi: 10.3389/fimmu.2018.01506. eCollection 2018. Front Immunol. 2018. PMID: 30013564 Free PMC article. - Promoter Proximity Defines Mutation Window for VH and VΚ Genes Rearranged to Different J Genes.
Heltzel JHM, Maul RW, Yang W, Gearhart PJ. Heltzel JHM, et al. J Immunol. 2022 May 1;208(9):2220-2226. doi: 10.4049/jimmunol.2101002. Epub 2022 Apr 13. J Immunol. 2022. PMID: 35418469 Free PMC article.
References
- Muramatsu, M., K. Kinoshita, S. Faragasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. - PubMed
- Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Piebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Lagelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell. 102:565–575. - PubMed
- Chaudhuri, J., M. Tian, C. Khuong, K. Chua, E. Pinaud, and F.W. Alt. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730. - PubMed
- Ramiro, A.R., P. Stavropoulos, M. Jankovic, and M.C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous