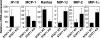Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis - PubMed (original) (raw)
Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis
Estelle Bettelli et al. J Exp Med. 2004.
Abstract
The transcription factors signal transducer and activator of transcription (STAT)1 and T-bet control the differentiation of interferon (IFN)-gamma-producing T helper type (Th)1 cells. Here we compare the role of T-bet and STAT1 in the initiation and regulation of experimental autoimmune encephalomyelitis (EAE), a disease initiated by Th1 cells. T-bet-deficient mice immunized with myelin oligodendrocyte glycoprotein (MOG) were resistant to the development of EAE. This protection was also observed when T-bet(-/-) mice were crossed to the MOG-specific 2D2 T cell receptor transgenic strain. In contrast, although T-bet is downstream of STAT1, STAT1(-/-) mice were highly susceptible to EAE and developed more severe and accelerated disease with atypical neuropathologic features. The function of T-bet was dominant as mice deficient in both T-bet and STAT1 were also protected from EAE. CD4(+) CD25(+) regulatory T cells from these two mice strains were fully competent and do not explain the difference in disease susceptibility. However, enhanced EAE in STAT1(-/-) mice was associated with continued generation of IFN-gamma-producing Th1 cells and up-regulation of selective chemokines responsible for the increased recruitment of macrophages and neutrophils in the central nervous system. Although the two transcription factors, STAT1 and T-bet, both induce IFN-gamma gene transcription, our results demonstrate marked differences in their function in regulating pathogenic Th1 cell responses.
Figures
Figure 1.
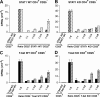
T-bet–deficient mice are resistant to EAE induction, whereas STAT1−/− mice develop fulminant EAE. (A) EAE development in T-bet– and STAT1-deficient mice. Groups of T-bet−/− and T-bet−/+ mice and STAT1−/− and 129 wild-type mice (6–7 mice per group) were immunized with MOG 35-55 and injected with pertussis toxin (a and c). T-bet−/− or T-bet+/+ 2D2 MOG-specific TCR transgenic mice were injected with pertussis toxin (b). Mice were observed daily for the development of EAE. (B) Histology of 2D2 TCR transgenic mice with spontaneous EAE. (a) Posterior spinal cord of a 2D2 × T-bet+/+ mouse with spontaneous EAE. There are small numbers of typical mononuclear cell perivascular infiltrates in leptomeninges and parenchyma (arrows). There were similar infiltrates in 2D2 × STAT1+/+ mice (not depicted). Magnification, 57. (b) Posterior spinal cord of 2D2 × T-bet−/− mouse. There are no inflammatory lesions. Magnification, 57. (c) Posterior spinal cord of 2D2 × STAT1−/− mouse with severe clinical disease. There is a large symmetric destructive inflammatory lesion in the posterior columns. Inset, higher power demonstrates confluent parenchymal inflammatory cell infiltration, predominantly by neutrophils. Magnification, 57; magnification of inset, 450. (d) Adjacent section to (c) demonstrating absence of axons in lesion center. Inset, higher power demonstrates neutrophils and complete loss of axons. Intact black fibers are axons on the periphery of the lesion. Magnification, 57; magnification of inset, 450. (e) Brain stem of 2D2 × STAT1−/− mouse with atypical disease. A large subpial/subependymal inflammatory lesion adjacent to the lateral foramen in the pons fills most of the field. Myelinated fibers (blue) are preserved in the lower left. *, cerebellum; magnification, 71. (f) Higher power of brain stem lesion in (e) with fibrin exudation from a vessel (arrow). Neutrophils and foamy macrophages are the predominant inflammatory cells. Magnification, 286. a–c, e, and f, Luxol fast blue/hematoxylin and eosin stain; d, Bielschowsky silver impregnation for axons.
Figure 1.
T-bet–deficient mice are resistant to EAE induction, whereas STAT1−/− mice develop fulminant EAE. (A) EAE development in T-bet– and STAT1-deficient mice. Groups of T-bet−/− and T-bet−/+ mice and STAT1−/− and 129 wild-type mice (6–7 mice per group) were immunized with MOG 35-55 and injected with pertussis toxin (a and c). T-bet−/− or T-bet+/+ 2D2 MOG-specific TCR transgenic mice were injected with pertussis toxin (b). Mice were observed daily for the development of EAE. (B) Histology of 2D2 TCR transgenic mice with spontaneous EAE. (a) Posterior spinal cord of a 2D2 × T-bet+/+ mouse with spontaneous EAE. There are small numbers of typical mononuclear cell perivascular infiltrates in leptomeninges and parenchyma (arrows). There were similar infiltrates in 2D2 × STAT1+/+ mice (not depicted). Magnification, 57. (b) Posterior spinal cord of 2D2 × T-bet−/− mouse. There are no inflammatory lesions. Magnification, 57. (c) Posterior spinal cord of 2D2 × STAT1−/− mouse with severe clinical disease. There is a large symmetric destructive inflammatory lesion in the posterior columns. Inset, higher power demonstrates confluent parenchymal inflammatory cell infiltration, predominantly by neutrophils. Magnification, 57; magnification of inset, 450. (d) Adjacent section to (c) demonstrating absence of axons in lesion center. Inset, higher power demonstrates neutrophils and complete loss of axons. Intact black fibers are axons on the periphery of the lesion. Magnification, 57; magnification of inset, 450. (e) Brain stem of 2D2 × STAT1−/− mouse with atypical disease. A large subpial/subependymal inflammatory lesion adjacent to the lateral foramen in the pons fills most of the field. Myelinated fibers (blue) are preserved in the lower left. *, cerebellum; magnification, 71. (f) Higher power of brain stem lesion in (e) with fibrin exudation from a vessel (arrow). Neutrophils and foamy macrophages are the predominant inflammatory cells. Magnification, 286. a–c, e, and f, Luxol fast blue/hematoxylin and eosin stain; d, Bielschowsky silver impregnation for axons.
Figure 2.
CD4+ CD25+ from both T-bet– and STAT1-deficient mice can suppress the proliferation of effector CD4+ T cells. CD4+ CD25+ and CD4+ CD25− T cells from STAT1−/−, T-bet−/−, and respective wild-type littermates were purified from the spleens and lymph nodes as described in Materials and Methods. Proliferation of CD4+ CD25− cells in the presence of irradiated APCs, 1 μg/ml anti-CD3 antibody, and different ratio of CD4+ CD25+ T cells (from STAT1 knockout, bold hatched bars; STAT1 wild-type, hatched bars; T-bet knockout, closed bars; T-bet wild-type, dotted bars) was determined by thymidine incorporation. These results are representative of four independent experiments.
Figure 3.
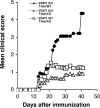
In absence of T-bet, STAT1−/− mice are protected from EAE. Groups of T-bet−/− (n = 14), STAT1−/− (n = 13), and STAT1−/− T-bet−/− (n = 13) mice were immunized with MOG 35-55 and injected with pertussis toxin. The development of EAE was followed over time. The data represent two independent experiments.
Figure 4.
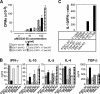
Proliferative response and cytokine production of T cells from T-bet– and STAT1-deficient mice. CD4+ T cells from naive 2D2 × STAT1−/−, 2D2 × T-bet−/−, 2D2 (T-bet × STAT1)−/− mice, and relative wild-type littermates were stimulated in vitro with MOG 35-55 peptide and syngeneic APCs. Proliferation was measured 72 h later by [3H]thymidine incorporation (A). The data are representative of at least four independent experiments. IFN-γ, IL−10, IL-5, IL-4, and TGF-β (B) produced in the culture supernatant was measured by ELISA 48 h after stimulation with peptide MOG 35-55 and APCs. (C) Ratio of IL-10/IFN-γ production in CD4+ T cells from 2D2 × T-bet−/−, 2D2 × T-bet+/+, 2D2 × STAT1−/−, 2D2 × STAT1+/+, 2D2 (T-bet × STAT1)−/−, and 2D2 (T-bet × STAT1)+/+. The data are presented as the average between three representative experiments ± SE.
Figure 5.
Chemokine expression in the brain of T-bet– and STAT1-deficient mice during EAE. Groups of STAT1−/− and STAT1+/+ mice (five mice/group) were immunized with MOG 35-55 and pertussis toxin. During the first attack of EAE, the brain from these mice was removed and chemokine expression was determined by real-time PCR. The results represent the mean relative mRNA expression (2-ΔΔCT × 1,000) of a particular chemokine within a group ± SE between samples of this group.
Similar articles
- T-bet is a STAT1-induced regulator of IL-12R expression in naïve CD4+ T cells.
Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. Afkarian M, et al. Nat Immunol. 2002 Jun;3(6):549-57. doi: 10.1038/ni794. Epub 2002 May 13. Nat Immunol. 2002. PMID: 12006974 - Functional and pathogenic differences of Th1 and Th17 cells in experimental autoimmune encephalomyelitis.
Domingues HS, Mues M, Lassmann H, Wekerle H, Krishnamoorthy G. Domingues HS, et al. PLoS One. 2010 Nov 29;5(11):e15531. doi: 10.1371/journal.pone.0015531. PLoS One. 2010. PMID: 21209700 Free PMC article. - T-bet is essential for the progression of experimental autoimmune encephalomyelitis.
Nath N, Prasad R, Giri S, Singh AK, Singh I. Nath N, et al. Immunology. 2006 Jul;118(3):384-91. doi: 10.1111/j.1365-2567.2006.02385.x. Immunology. 2006. PMID: 16827899 Free PMC article. - Cytokine shifts and tolerance in experimental autoimmune encephalomyelitis.
Chitnis T, Khoury SJ. Chitnis T, et al. Immunol Res. 2003;28(3):223-39. doi: 10.1385/IR:28:3:223. Immunol Res. 2003. PMID: 14713716 Review. - More than just a T-box: the role of T-bet as a possible biomarker and therapeutic target in autoimmune diseases.
Ji N, Sosa RA, Forsthuber TG. Ji N, et al. Immunotherapy. 2011 Mar;3(3):435-41. doi: 10.2217/imt.10.111. Immunotherapy. 2011. PMID: 21395384 Free PMC article. Review.
Cited by
- Improved insulin sensitivity despite increased visceral adiposity in mice deficient for the immune cell transcription factor T-bet.
Stolarczyk E, Vong CT, Perucha E, Jackson I, Cawthorne MA, Wargent ET, Powell N, Canavan JB, Lord GM, Howard JK. Stolarczyk E, et al. Cell Metab. 2013 Apr 2;17(4):520-33. doi: 10.1016/j.cmet.2013.02.019. Cell Metab. 2013. PMID: 23562076 Free PMC article. - Essential role of NK cells in IgG therapy for experimental autoimmune encephalomyelitis.
Chong WP, Ling MT, Liu Y, Caspi RR, Wong WM, Wu W, Tu W, Lau YL. Chong WP, et al. PLoS One. 2013;8(4):e60862. doi: 10.1371/journal.pone.0060862. Epub 2013 Apr 5. PLoS One. 2013. PMID: 23577171 Free PMC article. - Revisiting the old link between infection and autoimmune disease with commensals and T helper 17 cells.
Blander JM, Torchinsky MB, Campisi L. Blander JM, et al. Immunol Res. 2012 Dec;54(1-3):50-68. doi: 10.1007/s12026-012-8311-9. Immunol Res. 2012. PMID: 22460741 Free PMC article. Review. - Effector and regulatory T-cell subsets in autoimmunity and tissue inflammation.
Jäger A, Kuchroo VK. Jäger A, et al. Scand J Immunol. 2010 Sep;72(3):173-84. doi: 10.1111/j.1365-3083.2010.02432.x. Scand J Immunol. 2010. PMID: 20696013 Free PMC article. Review. - T-bet negatively regulates autoimmune myocarditis by suppressing local production of interleukin 17.
Rangachari M, Mauermann N, Marty RR, Dirnhofer S, Kurrer MO, Komnenovic V, Penninger JM, Eriksson U. Rangachari M, et al. J Exp Med. 2006 Aug 7;203(8):2009-19. doi: 10.1084/jem.20052222. Epub 2006 Jul 31. J Exp Med. 2006. PMID: 16880257 Free PMC article.
References
- Seder, R.A., and W.E. Paul. 1994. Acquisition of lymphokine-producing phenotype by CD4+ T cells. Annu. Rev. Immunol. 12:635–673. - PubMed
- Murphy, K.M., W. Ouyang, J.D. Farrar, J. Yang, S. Ranganath, H. Asnagli, M. Afkarian, and T.L. Murphy. 2000. Signaling and transcription in T helper development. Annu. Rev. Immunol. 18:451–494. - PubMed
- Szabo, S.J., S.T. Kim, G.L. Costa, X. Zhang, C.G. Fathman, and L.H. Glimcher. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 100:655–669. - PubMed
- O'Garra, A., and N. Arai. 2000. The molecular basis of T helper 1 and T helper 2 cell differentiation. Trends Cell Biol. 10:542–550. - PubMed
- Wekerle, H. 1993. Experimental autoimmune encephalomyelitis as a model of immune-mediated CNS disease. Curr. Opin. Neurobiol. 3:779–784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1P01 NS38037-04/NS/NINDS NIH HHS/United States
- 2P01 AI39671-07/AI/NIAID NIH HHS/United States
- 2R37 NS30843-11/NS/NINDS NIH HHS/United States
- 1R01 NS35685-06/NS/NINDS NIH HHS/United States
- 1R01 AI44880-03/AI/NIAID NIH HHS/United States
- AI48126/AI/NIAID NIH HHS/United States
- R01 AI044880/AI/NIAID NIH HHS/United States
- P01 AI039671/AI/NIAID NIH HHS/United States
- R37 NS030843/NS/NINDS NIH HHS/United States
- R01 NS035685/NS/NINDS NIH HHS/United States
- NS046414/NS/NINDS NIH HHS/United States
- R01 NS046414/NS/NINDS NIH HHS/United States
- P01 NS038037/NS/NINDS NIH HHS/United States
- R01 AI048126/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous