New real-time quantitative PCR procedure for quantification of bifidobacteria in human fecal samples - PubMed (original) (raw)
New real-time quantitative PCR procedure for quantification of bifidobacteria in human fecal samples
Miguel Gueimonde et al. Appl Environ Microbiol. 2004 Jul.
Abstract
The application of a real-time quantitative PCR method (5' nuclease assay), based on the use of a probe labeled at its 5' end with a stable, fluorescent lanthanide chelate, for the quantification of human fecal bifidobacteria was evaluated. The specificities of the primers and the primer-probe combination were evaluated by conventional PCR and real-time PCR, respectively. The results obtained by real-time PCR were compared with those obtained by fluorescent in situ hybridization, the current gold standard for intestinal microbiota quantification. In general, a good correlation between the two methods was observed. In order to determine the detection limit and the accuracy of the real-time PCR procedure, germfree rat feces were spiked with known amounts of bifidobacteria and analyzed by both methods. The detection limit of the method used in this study was found to be about 5 x 10(4) cells per g of feces. Both methods, real-time PCR and fluorescent in situ hybridization, led to an accurate quantification of the spiked samples with high levels of bifidobacteria, but real-time PCR was more accurate for samples with low levels. We conclude that the real-time PCR procedure described here is a specific, accurate, rapid, and easy method for the quantification of bifidobacteria in feces.
Figures
FIG. 1.
Calibration curve obtained by plotting C t values as a linear function of the base 10 logarithm of the initial number of bifidobacteria (B. infantis DSM 20088T, B. longum JCM 1217T, and B. adolescentis JCM 1275T) in the culture determined by plate counting.
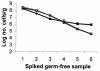
FIG. 2.
Results of the analysis of germfree rat feces spiked with known amounts of B. infantis DSM 20088. □, cells added; ○, cells detected by FISH; ×, cells detected by real-time PCR.
Similar articles
- Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula.
Haarman M, Knol J. Haarman M, et al. Appl Environ Microbiol. 2005 May;71(5):2318-24. doi: 10.1128/AEM.71.5.2318-2324.2005. Appl Environ Microbiol. 2005. PMID: 15870317 Free PMC article. Clinical Trial. - Quantitative assessment of faecal bifidobacterial populations by real-time PCR using lanthanide probes.
Gueimonde M, Debor L, Tölkkö S, Jokisalo E, Salminen S. Gueimonde M, et al. J Appl Microbiol. 2007 Apr;102(4):1116-22. doi: 10.1111/j.1365-2672.2006.03145.x. J Appl Microbiol. 2007. PMID: 17381755 - PCR and real-time PCR primers developed for detection and identification of Bifidobacterium thermophilum in faeces.
Mathys S, Lacroix C, Mini R, Meile L. Mathys S, et al. BMC Microbiol. 2008 Oct 10;8:179. doi: 10.1186/1471-2180-8-179. BMC Microbiol. 2008. PMID: 18847469 Free PMC article. - Genus- and species-specific PCR primers for the detection and identification of bifidobacteria.
Matsuki T, Watanabe K, Tanaka R. Matsuki T, et al. Curr Issues Intest Microbiol. 2003 Sep;4(2):61-9. Curr Issues Intest Microbiol. 2003. PMID: 14503690 Review. - [Quantitative PCR in the diagnosis of Leishmania].
Mortarino M, Franceschi A, Mancianti F, Bazzocchi C, Genchi C, Bandi C. Mortarino M, et al. Parassitologia. 2004 Jun;46(1-2):163-7. Parassitologia. 2004. PMID: 15305709 Review. Italian.
Cited by
- Microarray analysis reveals marked intestinal microbiota aberrancy in infants having eczema compared to healthy children in at-risk for atopic disease.
Nylund L, Satokari R, Nikkilä J, Rajilić-Stojanović M, Kalliomäki M, Isolauri E, Salminen S, de Vos WM. Nylund L, et al. BMC Microbiol. 2013 Jan 23;13:12. doi: 10.1186/1471-2180-13-12. BMC Microbiol. 2013. PMID: 23339708 Free PMC article. - Poor Bifidobacterial Colonization Is Associated with Late Provision of Colostrum and Improved with Probiotic Supplementation in Low Birth Weight Infants.
Tanaka K, Nakamura Y, Terahara M, Yanagi T, Nakahara S, Furukawa O, Tsutsui H, Inoue R, Tsukahara T, Koshida S. Tanaka K, et al. Nutrients. 2019 Apr 13;11(4):839. doi: 10.3390/nu11040839. Nutrients. 2019. PMID: 31013872 Free PMC article. Clinical Trial. - Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly.
Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Collado MC, et al. Appl Environ Microbiol. 2007 Dec;73(23):7767-70. doi: 10.1128/AEM.01477-07. Epub 2007 Oct 12. Appl Environ Microbiol. 2007. PMID: 17933936 Free PMC article. - Comparison of four polymerase chain reaction methods for the rapid detection of human fecal pollution in marine and inland waters.
Bachoon DS, Miller CM, Green CP, Otero E. Bachoon DS, et al. Int J Microbiol. 2010;2010:595692. doi: 10.1155/2010/595692. Epub 2010 Aug 5. Int J Microbiol. 2010. PMID: 20811614 Free PMC article. - Establishment of an analytical system for the human fecal microbiota, based on reverse transcription-quantitative PCR targeting of multicopy rRNA molecules.
Matsuda K, Tsuji H, Asahara T, Matsumoto K, Takada T, Nomoto K. Matsuda K, et al. Appl Environ Microbiol. 2009 Apr;75(7):1961-9. doi: 10.1128/AEM.01843-08. Epub 2009 Feb 5. Appl Environ Microbiol. 2009. PMID: 19201979 Free PMC article.
References
- Benno, Y., and T. Mitsuoka. 1986. Development of intestinal microflora in humans and animals. Bifidobateria Microflora 5:13-25.
- Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical