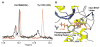Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains - PubMed (original) (raw)
Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains
Maria Victoria E Botuyan et al. Structure. 2004 Jul.
Abstract
BRCT tandem domains, found in many proteins involved in DNA damage checkpoint and DNA repair pathways, were recently shown to be phosphopeptide binding motifs. Using solution nuclear magnetic resonance (NMR) spectroscopy and mutational analysis, we have characterized the interaction of BRCA1-BRCT domains with a phosphoserine-containing peptide derived from the DNA repair helicase BACH1. We show that a phenylalanine in the +3 position from the phosphoserine of BACH1 is bound to a conserved hydrophobic pocket formed between the two BRCT domains and that recognition of the phosphate group is mediated by lysine and serine side chains from the amino-terminal BRCT domain. Mutations that prevent phosphopeptide binding abolish BRCA1 function in DNA damage-induced checkpoint control. Our NMR data also reveal a dynamic interaction between BRCA1-BRCT and BACH1, where the bound phosphopeptide exists as an equilibrium of two conformations and where BRCA1-BRCT undergoes a transition to a more rigid conformation upon peptide binding.
Figures
Figure 1. Amino Acid Sequence of BACH1 Phosphopeptide
The numbering scheme adopted for BACH1 phosphopeptide (BACH1p) is 1 to 13. The sequence corresponds to residues 985–998 of BACH1 protein. The phosphorylated Ser is indicated by pSer6. Relative positions of other residues from pSer6 are also indicated.
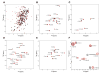
Figure 2. Variation in BRCA1-BRCT Chemical Shifts upon BACH1p Binding
(A) Overlay of 15N-HSQC spectra of 15N/[50%2H]-labeled BRCA1-BRCT, free (black) and in a 1:1 complex with nonlabeled BACH1p (red), illustrating the extent of chemical shift perturbations upon peptide binding. (B–E) Overlay of 15N-HSQC spectra of BRCA1-BRCT, free (black) and bound to nonlabeled BACH1p (red), selectively 15N-labeled for Leu, Tyr, Val, and Phe, respectively. (F) Overlay of a selected region of the 13C-HSQC spectrum of 15N/13C/2H-labeled BRCA1-BRCT, but protonated at the methyl groups of Val, Leu, and Ile (δ1), in the absence (black) and presence (red) of nonlabeled BACH1p.
Figure 3. Summary of NMR Data for BRCA1-BRCT in Complex with BACH1p
(A) Sequence of BRCA1-BRCT (1646–1859) with a summary of secondary structure elements derived from NMR data of the BACH1p-bound protein: interproton NOEs, solvent protection, TALOS, and chemical shift index (CSI) results. NOEs were measured from 100, 200, and 300 ms mixing times 3D 15N-edited and 13C-edited NOESY data recorded on 50%, 60%, and 100% deuterated 15N- and 15N/13C-labeled BRCA1-BRCT samples, as well as on a 15N/13C-labeled and deuterated (but protonated at Val, Leu, and Ile δ1 methyls) protein. An NOE is indicated by a bar; the thickness is proportional to intensity. A solid circle represents a residue significantly protected from 2H exchange (i.e., 1H signal is still visible in the HSQC spectrum 4 hr after exchange in D2O). Solid and empty squares indicate that Phi and Psi angles predicted by TALOS are compatible with those expected for α helices and β strands, respectively. Negative and positive CSI are designated by a square below and above the line, respectively. β strands (arrows) and α helices (coils) deduced from all the NMR data are summarized (NMR). Thin lines connecting the β strands indicate that long-range HNi-HNj and Hαi-HNj NOEs have been observed. The NMR data are in very good agreement with the secondary structures found in the crystal structure of free BRCA1-BRCT (X-ray). (B) Mapping of interresidue NOEs on the crystal structure of free BRCA1-BRCT (PDB entry 1JNX). Protons involved in NOEs are connected by red lines; 453 medium-range and 238 long-range NOEs are shown. (C) Residues involved in intermolecular NOEs are connected by red double arrows.
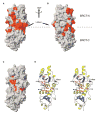
Figure 4. Identification of the BACH1p Binding Site on BRCA1-BRCT
(A) Changes in chemical shifts mapped on the crystal structure of free BRCA-BRCT. Shown in red are residues that are affected upon BACH1p binding. (B) Same as (A) after 180° rotation about the vertical axis. (C) Residues involved in intermolecular NOEs with BACH1p mapped on the crystal structure of free BRCA-BRCT. The region involved in phosphoserine binding is circled in yellow, while the hydrophobic pocket is indicated by a red circle. (D) Stereo ribbon representation of BRCA1-BRCT showing the amino acids involved in intermolecular NOEs, as well as Met1775 and Lys1702. Red and yellow circles are defined as in (C).
Figure 5. Evidence of Conformational Equilibrium in Bound BACH1p
Regions from the 2D 1H-1H NOESY (A), F1/F2 double-filtered NOESY (B), F2-filtered NOESY (C), and F2 15N-edited NOESY (D) spectra of the free BACH1p peptide (A) and BRCA1-BRCT/BACH1p complex (B–D). BACH1p peptide was nonlabeled in (A) to (D). BRCA1-BRCT was 15N/13C/2H-labeled with protonated methyls (Val, Leu, and Ile δ1) in (B) and (C) and 15N/13C/[60%-2H]-labeled in (D). The F2 dimension in (A)–(C) corresponds to the aromatic region of BACH1p Phe9. The F2 dimension in (D) corresponds to the HN of Thr1700 without 15N decoupling. Experiments were done in D2O at 30°C and pH 7.5. Thr1700 HN is protected from exchange with D2O as seen in (D). (A) and (B) show intrapeptide NOEs. (C) displays both intrapeptide and peptide/protein NOEs. (D) contains intraprotein and peptide/protein NOEs. Note two sets of signals for Phe9 Hα, Thr8 Hβ, and Thr8 Hγ indicating two conformations of bound BACH1p in (B)–(D). In one of these conformations, Phe 9 is close in space to Pro7, as indicated by a box in (B) and (C).
Figure 6. Tyr1703 Connnects the Phophopeptide Binding Site to the Inter-BRCT Linker
(A) Superposition of a selected region from the 1H spectra of 15N/13C/2H-labeled BRCA1-BRCT, free (black) and bound to nonlabeled BACH1p (red), showing the hydroxyl proton of Tyr1703. Note that other 1H signals in the spectra are 15N-coupled. Experiments were recorded at 30°C and pH 7.5. (B) Location of Tyr1703 in the 3D structure of BRCA1-BRCT. Tyr1703 bridges the phosphopeptide binding site to the inter-BRCT linker region.
Figure 7. S1655A and K1702A Mutants of BRCA1 Do Not Interact with BACH1 In Vitro and In Vivo
(A) 293T cell lysates were incubated with wild-type or mutant GST-BRCA1-BRCT proteins immobilized on Sepharose beads. Proteins bound to beads were eluted and separated by SDS-PAGE. Western blots were performed with anti-BACH1 antibody (upper panel). Western blots of whole-cell lysates were also included to demonstrate equal amounts of BACH1 in these samples (lower panel). (B) Myc epitope-tagged wild-type and mutant BRCA1 were transiently expressed in 293T cells. Immunoprecipitation and immunoblotting experiments were performed with anti-myc or anti-BACH1 antibodies as indicated.
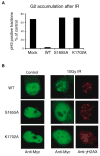
Figure 8. Phosphoprotein Binding Activity of BRCA1 Is Required for DNA Damage-Induced G2/M Checkpoint Control and BRCA1 Nuclear Foci Formation
Wild-type or mutant BRCA1 was transiently expressed in HCC1937 cells. (A) G2/M checkpoint assays were conducted as described in the Experimental Procedures. The fraction of cells in M phase is expressed as a percentage of that measured in unirradiated control cells. (B) Three hours after γ-irradiation, immunostaining experiments were performed with the indicated antibodies.
Similar articles
- Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains.
Rodriguez M, Yu X, Chen J, Songyang Z. Rodriguez M, et al. J Biol Chem. 2003 Dec 26;278(52):52914-8. doi: 10.1074/jbc.C300407200. Epub 2003 Oct 24. J Biol Chem. 2003. PMID: 14578343 - Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling.
Shiozaki EN, Gu L, Yan N, Shi Y. Shiozaki EN, et al. Mol Cell. 2004 May 7;14(3):405-12. doi: 10.1016/s1097-2765(04)00238-2. Mol Cell. 2004. PMID: 15125843 - Structural basis for cell cycle checkpoint control by the BRCA1-CtIP complex.
Varma AK, Brown RS, Birrane G, Ladias JA. Varma AK, et al. Biochemistry. 2005 Aug 23;44(33):10941-6. doi: 10.1021/bi0509651. Biochemistry. 2005. PMID: 16101277 - Phosphopeptide interactions with BRCA1 BRCT domains: More than just a motif.
Wu Q, Jubb H, Blundell TL. Wu Q, et al. Prog Biophys Mol Biol. 2015 Mar;117(2-3):143-148. doi: 10.1016/j.pbiomolbio.2015.02.003. Epub 2015 Feb 17. Prog Biophys Mol Biol. 2015. PMID: 25701377 Free PMC article. Review. - Cancer predisposing mutations in BRCT domains.
di Masi A, Gullotta F, Cappadonna V, Leboffe L, Ascenzi P. di Masi A, et al. IUBMB Life. 2011 Jul;63(7):503-12. doi: 10.1002/iub.472. IUBMB Life. 2011. PMID: 21698754 Review.
Cited by
- Spectrum of BRCA1 interacting helicase 1 aberrations and potential prognostic and therapeutic implication: a pan cancer analysis.
Long G, Hu K, Zhang X, Zhou L, Li J. Long G, et al. Sci Rep. 2023 Mar 17;13(1):4435. doi: 10.1038/s41598-023-31109-6. Sci Rep. 2023. PMID: 36932143 Free PMC article. - NBS1 binds directly to TOPBP1 via disparate interactions between the NBS1 BRCT1 domain and the TOPBP1 BRCT1 and BRCT2 domains.
Huynh O, Ruis K, Montales K, Michael WM. Huynh O, et al. DNA Repair (Amst). 2023 Mar;123:103461. doi: 10.1016/j.dnarep.2023.103461. Epub 2023 Feb 2. DNA Repair (Amst). 2023. PMID: 36738687 Free PMC article. - Wwox Binding to the Murine Brca1-BRCT Domain Regulates Timing of Brip1 and CtIP Phospho-Protein Interactions with This Domain at DNA Double-Strand Breaks, and Repair Pathway Choice.
Park D, Gharghabi M, Reczek CR, Plow R, Yungvirt C, Aldaz CM, Huebner K. Park D, et al. Int J Mol Sci. 2022 Mar 28;23(7):3729. doi: 10.3390/ijms23073729. Int J Mol Sci. 2022. PMID: 35409089 Free PMC article. - Pre-ribosomal RNA reorganizes DNA damage repair factors in nucleus during meiotic prophase and DNA damage response.
Gai X, Xin D, Wu D, Wang X, Chen L, Wang Y, Ma K, Li Q, Li P, Yu X. Gai X, et al. Cell Res. 2022 Mar;32(3):254-268. doi: 10.1038/s41422-021-00597-4. Epub 2022 Jan 4. Cell Res. 2022. PMID: 34980897 Free PMC article. - Structural basis of homologous recombination.
Sun Y, McCorvie TJ, Yates LA, Zhang X. Sun Y, et al. Cell Mol Life Sci. 2020 Jan;77(1):3-18. doi: 10.1007/s00018-019-03365-1. Epub 2019 Nov 20. Cell Mol Life Sci. 2020. PMID: 31748913 Free PMC article. Review.
References
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. - PubMed
- Botuyan MVE, Mer G, Yi GS, Koth CM, Case DA, Edwards AM, Chazin WJ, Arrowsmith CH. Solution structure and dynamics of yeast elongin C in complex with a von Hippel-Lindau peptide. J Mol Biol. 2001;312:177–186. - PubMed
- Caldecott KW. The BRCT domain: signaling with friends? Science. 2003;302:579–580. - PubMed
- Callebaut I, Mornon JP. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. - PubMed
- Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous