The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins - PubMed (original) (raw)
. 2004 Jul 19;166(2):213-23.
doi: 10.1083/jcb.200403069. Epub 2004 Jul 12.
Greg M Findlay, Alex Gray, Tatiana Tolkacheva, Simon Wigfield, Heike Rebholz, Jill Barnett, Nick R Leslie, Susan Cheng, Peter R Shepherd, Ivan Gout, C Peter Downes, Richard F Lamb
Affiliations
- PMID: 15249583
- PMCID: PMC2172316
- DOI: 10.1083/jcb.200403069
The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins
Laura S Harrington et al. J Cell Biol. 2004.
Abstract
Insulin-like growth factors elicit many responses through activation of phosphoinositide 3-OH kinase (PI3K). The tuberous sclerosis complex (TSC1-2) suppresses cell growth by negatively regulating a protein kinase, p70S6K (S6K1), which generally requires PI3K signals for its activation. Here, we show that TSC1-2 is required for insulin signaling to PI3K. TSC1-2 maintains insulin signaling to PI3K by restraining the activity of S6K, which when activated inactivates insulin receptor substrate (IRS) function, via repression of IRS-1 gene expression and via direct phosphorylation of IRS-1. Our results argue that the low malignant potential of tumors arising from TSC1-2 dysfunction may be explained by the failure of TSC mutant cells to activate PI3K and its downstream effectors.
Copyright The Rockerfeller University Press
Figures
Figure 1.
PI3K activation by insulin-like growth factors is dependent on functional TSC2 . (A) PKB activation in cells expressing or lacking functional TSC2. PKB activation in isogenic TSC2 +/+, TSC2 −/− MEFs, and TSC2 −/− MEF lines in which wild-type (−/− (+WT)) or a pathogenic mutant TSC2 (−/− (+N1643K)) has been reintroduced were starved and stimulated with the indicated growth factors for 10 min. The ability of insulin, IGF-1, and EGF to activate PKB is demonstrated by phosphorylation of Ser-473 (S473-P). (B) Phosphorylation of GSK3α/β, at GSK3α Ser-21 (top) and GSK3β Ser-9 (middle) after insulin, IGF-1, or EGF stimulation. Bottom: total GSK3α/β. (C) PI3K activity in TSC2 +/+ or TSC2 −/− MEFs, or TSC2 −/− MEFs reconstituted with wild-type TSC2 (+WT) or a disease-causing mutant TSC2 (+N1643K) after stimulation with IGF-1. Results are expressed as values relative to unstimulated controls. (D) PIP3 levels in TSC2 +/+ or TSC2 −/− MEFs, or TSC2 −/− MEFs reconstituted with wild-type TSC2 (+WT) or a disease-causing mutant TSC2 (+N1643K) after stimulation with IGF-1 or EGF. The results (means and SD of triplicate determinations) are expressed relative to control unstimulated cells.
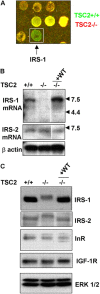
Figure 2.
IRS-1 mRNA and protein expression is dependent on TSC2. (A) Microarray analysis of TSC2 +/+ and TSC2 −/− MEFs. IRS-1 mRNA is more abundant in TSC2 +/+ MEFs (green) compared with TSC2 −/− MEFs (red). (B) Northern blotting of IRS-1 (top) or IRS-2 (middle) mRNAs in isogenic TSC2 +/+, TSC2 −/− MEFs, and a TSC2 −/− MEF line in which wild-type TSC2 has been reintroduced (−/− +WT). Aliquots of these samples were run on a separate gel and probed for β-actin as a control for RNA content (bottom). White line indicates that intervening lanes have been removed. (C) IRS-1 (first panel), IRS-2 (second panel), InR (third panel), and IGF1-R (fourth panel) protein levels in TSC2 +/+, TSC2 −/−, and a TSC2 −/− MEF line in which wild-type TSC2 has been reintroduced (−/− +WT). A blot was reprobed for ERK1/2 as a control for protein loading (bottom panel).
Figure 3.
Inhibition of S6K restores IRS-1 mRNA. (A) IRS-1 mRNA levels (top) in serum-starved TSC2 +/+ or TSC2 −/− MEFs in untreated cells or after the addition of 20 nM rapamycin for various times. The blot was reprobed for β-actin as a control for mRNA loading (bottom). White line indicates that intervening lanes have been removed. (B) IRS-1 mRNA levels (top) in TSC2 −/− MEFs treated with 20 nM rapamycin alone, or in the presence of 10 μg/ml actinomycin D for the final 10 h of a 24-h treatment. The blot was reprobed for β-actin as a control for mRNA loading (bottom). (C) Quantitative RT-PCR analysis of untreated, rapamycin-treated, and rapamycin plus actinomycin D–treated TSC2 −/− MEFs. Left, β-actin; right, IRS-1. A single graph from triplicate determinations showing identical results is shown. (D) RNAi-mediated inhibition of S6K1 and S6K2. Western blotting of extracts from TSC2 +/+ or TSC2 −/− MEFs transfected with a combination of scrambled siRNAs (C) or S6K1, S6K2, or S6K1 plus S6K2 siRNAs. Top, S6K1; middle, S6K2; bottom, anti-pS6 (Ser240/244). Where indicated, cells were stimulated for 10 min with insulin or treated with 20 nM rapamycin for 1 h. (E) IRS-1 mRNA levels in TSC2 +/+ MEFs or TSC2 −/− MEFs transfected with a combination of S6K1 and S6K2 scrambled siRNAs (C), S6K1, or S6K2 siRNAs, or treated for 24 h with 20 nM rapamycin. Top, IRS-1 mRNA; The blot was also reprobed for β-actin as a control for mRNA loading (bottom). (F) Quantitative RT-PCR analysis of untreated TSC2 +/+, TSC2 −/−, or TSC2 −/− treated with 20 nM rapamycin or RNAi to S6K1 or S6K2. Left, β-actin; right, IRS-1. A single graph from triplicate determinations showing identical results is shown.
Figure 3.
Inhibition of S6K restores IRS-1 mRNA. (A) IRS-1 mRNA levels (top) in serum-starved TSC2 +/+ or TSC2 −/− MEFs in untreated cells or after the addition of 20 nM rapamycin for various times. The blot was reprobed for β-actin as a control for mRNA loading (bottom). White line indicates that intervening lanes have been removed. (B) IRS-1 mRNA levels (top) in TSC2 −/− MEFs treated with 20 nM rapamycin alone, or in the presence of 10 μg/ml actinomycin D for the final 10 h of a 24-h treatment. The blot was reprobed for β-actin as a control for mRNA loading (bottom). (C) Quantitative RT-PCR analysis of untreated, rapamycin-treated, and rapamycin plus actinomycin D–treated TSC2 −/− MEFs. Left, β-actin; right, IRS-1. A single graph from triplicate determinations showing identical results is shown. (D) RNAi-mediated inhibition of S6K1 and S6K2. Western blotting of extracts from TSC2 +/+ or TSC2 −/− MEFs transfected with a combination of scrambled siRNAs (C) or S6K1, S6K2, or S6K1 plus S6K2 siRNAs. Top, S6K1; middle, S6K2; bottom, anti-pS6 (Ser240/244). Where indicated, cells were stimulated for 10 min with insulin or treated with 20 nM rapamycin for 1 h. (E) IRS-1 mRNA levels in TSC2 +/+ MEFs or TSC2 −/− MEFs transfected with a combination of S6K1 and S6K2 scrambled siRNAs (C), S6K1, or S6K2 siRNAs, or treated for 24 h with 20 nM rapamycin. Top, IRS-1 mRNA; The blot was also reprobed for β-actin as a control for mRNA loading (bottom). (F) Quantitative RT-PCR analysis of untreated TSC2 +/+, TSC2 −/−, or TSC2 −/− treated with 20 nM rapamycin or RNAi to S6K1 or S6K2. Left, β-actin; right, IRS-1. A single graph from triplicate determinations showing identical results is shown.
Figure 4.
Inhibition of S6K restores insulin signaling. (A) Treatment of IRS-1 immunoprecipitates with λ-phosphatase indicating that reduced migration of IRS-1 in TSC2 −/− MEFs is due to increased phosphorylation. (B) Treatment of serum-starved TSC2 +/+ or TSC2 −/− MEFs for various times with rapamycin followed by immunoblotting for IRS-1 (top) or IRS-2 (bottom). The slower migrating, more heavily phosphorylated IRS-1 or IRS-2 present in TSC2 −/− MEFs is indicated with an arrow; the faster migrating IRS-1 or IRS-2 present in TSC2 +/+ cells or after rapamycin treatment is indicated with an arrowhead. (C) PKB activation by insulin or IGF-1 after treatment of TSC2 +/+ or TSC2 −/− MEFs with rapamycin for the indicated times. Top, Ser473-phosphorylated PKB; bottom, level of total PKB. (D) PIP3 levels in TSC2 +/+ (gray bars) or TSC2 −/− (white bars) MEFs after IGF-1 stimulation in untreated cells or cells pretreated for 24 h with 20 nM rapamycin. Results from triplicate determinations (means and SDs) are shown normalized to control unstimulated samples and are representative of three independent experiments. (E) PKB activation after RNAi-mediated inhibition of IRS-1, IRS-2, IRS-1 plus IRS-2, or a combination of scrambled IRS-1 and IRS-2 siRNA (Scrambled) in TSC2 −/− MEFs stimulated by insulin for 10 min after treatment with 20 nM rapamycin for 1 h. First panel, IRS-1; second panel, IRS-2; third panel, Ser473-phosphorylated PKB; bottom panel, total PKB. (F) Rescue of PKB activation in TSC2 −/− MEFs after overexpression of IRS-1 or IRS-2. Extracts from mock-transfected TSC2 −/− MEFs (C) or MEFs transfected with expression constructs expressing myc-IRS-1 or IRS-2 were starved for 24 h after transfection and were treated with 20 nM rapamycin for 1 h before stimulation with insulin for 10 min. Where indicated, mock-transfected cells were also treated with 20 nM rapamycin for 24 h before stimulation. First panel, myc-IRS-1 detected with 9E10 monoclonal; second panel, IRS-2; third panel, Ser473-phosphorylated PKB; bottom panel, total PKB. (G) PKB activation in TSC2 −/− MEFs after RNAi-mediated inhibition of S6K1, S6K2, S6K1 plus S6K2, or a mixture of S6K1 and S6K2 scrambled siRNAs (Scrambled) after insulin stimulation for 10 min. Scrambled siRNA-transfected cells were also treated with 20 nM rapamycin for 24 h before insulin stimulation where indicated.
Figure 4.
Inhibition of S6K restores insulin signaling. (A) Treatment of IRS-1 immunoprecipitates with λ-phosphatase indicating that reduced migration of IRS-1 in TSC2 −/− MEFs is due to increased phosphorylation. (B) Treatment of serum-starved TSC2 +/+ or TSC2 −/− MEFs for various times with rapamycin followed by immunoblotting for IRS-1 (top) or IRS-2 (bottom). The slower migrating, more heavily phosphorylated IRS-1 or IRS-2 present in TSC2 −/− MEFs is indicated with an arrow; the faster migrating IRS-1 or IRS-2 present in TSC2 +/+ cells or after rapamycin treatment is indicated with an arrowhead. (C) PKB activation by insulin or IGF-1 after treatment of TSC2 +/+ or TSC2 −/− MEFs with rapamycin for the indicated times. Top, Ser473-phosphorylated PKB; bottom, level of total PKB. (D) PIP3 levels in TSC2 +/+ (gray bars) or TSC2 −/− (white bars) MEFs after IGF-1 stimulation in untreated cells or cells pretreated for 24 h with 20 nM rapamycin. Results from triplicate determinations (means and SDs) are shown normalized to control unstimulated samples and are representative of three independent experiments. (E) PKB activation after RNAi-mediated inhibition of IRS-1, IRS-2, IRS-1 plus IRS-2, or a combination of scrambled IRS-1 and IRS-2 siRNA (Scrambled) in TSC2 −/− MEFs stimulated by insulin for 10 min after treatment with 20 nM rapamycin for 1 h. First panel, IRS-1; second panel, IRS-2; third panel, Ser473-phosphorylated PKB; bottom panel, total PKB. (F) Rescue of PKB activation in TSC2 −/− MEFs after overexpression of IRS-1 or IRS-2. Extracts from mock-transfected TSC2 −/− MEFs (C) or MEFs transfected with expression constructs expressing myc-IRS-1 or IRS-2 were starved for 24 h after transfection and were treated with 20 nM rapamycin for 1 h before stimulation with insulin for 10 min. Where indicated, mock-transfected cells were also treated with 20 nM rapamycin for 24 h before stimulation. First panel, myc-IRS-1 detected with 9E10 monoclonal; second panel, IRS-2; third panel, Ser473-phosphorylated PKB; bottom panel, total PKB. (G) PKB activation in TSC2 −/− MEFs after RNAi-mediated inhibition of S6K1, S6K2, S6K1 plus S6K2, or a mixture of S6K1 and S6K2 scrambled siRNAs (Scrambled) after insulin stimulation for 10 min. Scrambled siRNA-transfected cells were also treated with 20 nM rapamycin for 24 h before insulin stimulation where indicated.
Figure 5.
IRS-1 is a novel S6K substrate. (A) Domain structure of IRS-1 indicating regions of GST fusions (aa numbering based on murine IRS-1) used for in vitro kinase assays and comparison of the S6 phosphorylation site with candidate S6-like phosphorylation sites present in IRS-1108–516. Conserved arginines are shown in bold, serines phosphorylated by S6K in S6 and potentially phosphorylated in IRS-1108–516 are shown in red, and serines mutated to alanines in mutants of sites 1–3 are underlined. Also shown is the homologous site to site 2 in murine IRS-2. (B) In vitro kinase assay of GST-IRS-1 fragments with purified S6K2. Full-length GST fusion proteins (1–4, as indicated in A) and 40S ribosomal proteins containing S6 (40S) in these preparations are indicated with asterisks in the Coomassie blue–stained gel (right), and the 32P-labeled proteins after the kinase assay and autoradiography are shown on the left. Two phosphorylated bands are noted in kinase assay with IRS-1108–516, the lower of which represents a poorly resolved doublet of cleaved forms of the protein. An arrow indicates 32P-labeled S6 present in the 40S ribosome preparation. (C) In vitro kinase assay of IRS-1 site-directed mutants by S6K1. Top panels, autoradiographs; bottom panels, Coomassie blue–stained gels; left panels, phosphorylation of IRS-1108–516 (WT) and site 1–3 compound mutants as indicated in Fig. 4 A; right panels, phosphorylation of IRS-1108–516 and individual serine-to-alanine mutants of the serine residues within site 2. (D) Ser302 phosphorylation of IRS-1108–516 wild-type (WT) or IRS-1108–516 with Ser302 mutated to alanine (S302A) detected with a phosphospecific antibody to IRS-1 Ser302 after in vitro phosphorylation by S6K1. (E) IRS-1 Ser302 phosphorylation is elevated in vivo. Top, Ser302-phosphorylated IRS-1 detected by a phosphospecific Ser302 antibody in extracts from serum-starved isogenic TSC2 +/+ and TSC2 −/− MEFs, and TSC2 −/− MEFs lines in which wild-type (−/− (+WT)) or a pathogenic mutant TSC2 (−/− (+N1643K)) has been reintroduced; top, Ser302-phosphorylated IRS-1; bottom, total IRS-1. (F) IRS-1 Ser302 phosphorylation is inhibited by mTOR/S6K inhibition with rapamycin in serum-starved TSC2 −/− MEFs. Top, Ser302-phosphorylated IRS-1; bottom, total IRS-1. Where indicated, MEFs were also stimulated with insulin for 10 min. (G) IRS-1 Ser302 phosphorylation is diminished in TSC2 −/− MEFs after siRNA-mediated inhibition of S6K1. Top, Ser302-phosphorylated IRS-1; bottom, total IRS-1. Where indicated, MEFs were also stimulated with insulin for 10 min.
Figure 5.
IRS-1 is a novel S6K substrate. (A) Domain structure of IRS-1 indicating regions of GST fusions (aa numbering based on murine IRS-1) used for in vitro kinase assays and comparison of the S6 phosphorylation site with candidate S6-like phosphorylation sites present in IRS-1108–516. Conserved arginines are shown in bold, serines phosphorylated by S6K in S6 and potentially phosphorylated in IRS-1108–516 are shown in red, and serines mutated to alanines in mutants of sites 1–3 are underlined. Also shown is the homologous site to site 2 in murine IRS-2. (B) In vitro kinase assay of GST-IRS-1 fragments with purified S6K2. Full-length GST fusion proteins (1–4, as indicated in A) and 40S ribosomal proteins containing S6 (40S) in these preparations are indicated with asterisks in the Coomassie blue–stained gel (right), and the 32P-labeled proteins after the kinase assay and autoradiography are shown on the left. Two phosphorylated bands are noted in kinase assay with IRS-1108–516, the lower of which represents a poorly resolved doublet of cleaved forms of the protein. An arrow indicates 32P-labeled S6 present in the 40S ribosome preparation. (C) In vitro kinase assay of IRS-1 site-directed mutants by S6K1. Top panels, autoradiographs; bottom panels, Coomassie blue–stained gels; left panels, phosphorylation of IRS-1108–516 (WT) and site 1–3 compound mutants as indicated in Fig. 4 A; right panels, phosphorylation of IRS-1108–516 and individual serine-to-alanine mutants of the serine residues within site 2. (D) Ser302 phosphorylation of IRS-1108–516 wild-type (WT) or IRS-1108–516 with Ser302 mutated to alanine (S302A) detected with a phosphospecific antibody to IRS-1 Ser302 after in vitro phosphorylation by S6K1. (E) IRS-1 Ser302 phosphorylation is elevated in vivo. Top, Ser302-phosphorylated IRS-1 detected by a phosphospecific Ser302 antibody in extracts from serum-starved isogenic TSC2 +/+ and TSC2 −/− MEFs, and TSC2 −/− MEFs lines in which wild-type (−/− (+WT)) or a pathogenic mutant TSC2 (−/− (+N1643K)) has been reintroduced; top, Ser302-phosphorylated IRS-1; bottom, total IRS-1. (F) IRS-1 Ser302 phosphorylation is inhibited by mTOR/S6K inhibition with rapamycin in serum-starved TSC2 −/− MEFs. Top, Ser302-phosphorylated IRS-1; bottom, total IRS-1. Where indicated, MEFs were also stimulated with insulin for 10 min. (G) IRS-1 Ser302 phosphorylation is diminished in TSC2 −/− MEFs after siRNA-mediated inhibition of S6K1. Top, Ser302-phosphorylated IRS-1; bottom, total IRS-1. Where indicated, MEFs were also stimulated with insulin for 10 min.
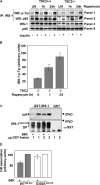
Figure 6.
S6K phosphorylation blocks IRS-1 function. (A) IRS-1 tyrosine phosphorylation and association with PI3K is suppressed in TSC2 −/− MEFs and rescued by inhibition of S6K. IRS-1 immunoprecipitates in unstimulated or insulin-stimulated TSC2 +/+ or TSC2 −/− MEFs after rapamycin treatment for the indicated times, immunoblotted for phosphotyrosine (panel 1), or the coimmunoprecipitation of the p85 subunit of PI3K (panel 2). The total levels of IRS-1 and p85 in cell lysates are indicated in panels 3 and 4, respectively. (B) Quantitation of insulin- stimulated IRS-1 tyrosine phosphorylation in untreated TSC2 −/− MEFs or the same cells after rapamycin treatment for the indicated times. An arbitrary value was obtained representing the difference between unstimulated and the corresponding insulin-stimulated level of phosphotyrosine in IRS-1 immunoprecipitations as detected on scanned autoradiographs. The values from three independent experiments were averaged and plotted on a bar chart, with the error bars showing the SEM. (C) Phosphorylation of immobilized GST-IRS-1108–516 PTB domain by S6K2 inhibits insulin receptor (InR) interaction. The interaction with tyrosine-phosphorylated InR (top) was determined in vitro by the addition of glutathione beads containing the indicated amounts of S6K2-phosphorylated or unphosphorylated GST-IRS-1108–516 or GST to lysates prepared from insulin-stimulated CHO-IR cells. An immunoblot of the level of GST-IRS-1 fusion protein or GST is indicated on the bottom. DP indicates degradation products of the GST-IRS-1 protein. (D) Quantitation of InR binding to GST-IRS-1108–516 (WT) or a mutant GST-IRS-1108–516 in which serine 302 is mutated to alanine (S302A) after in vitro phosphorylation by S6K2. Error bars show the SEM from three experiments. (E) InR binding of wild-type GST-IRS-1108–516, or S302A and S302E mutants in a pull-down assay. Top, InR; bottom, anti-GST. (F) Quantitation of InR binding to GST-IRS-1108–516 (WT) or S302A and S302E mutants. An arbitrary value of 100 was assigned for the binding of wild-type GST-IRS-1108–516, and the binding of the mutant IRS-1 proteins was expressed relative to this value. Error bars show the SEM from three experiments.
Figure 6.
S6K phosphorylation blocks IRS-1 function. (A) IRS-1 tyrosine phosphorylation and association with PI3K is suppressed in TSC2 −/− MEFs and rescued by inhibition of S6K. IRS-1 immunoprecipitates in unstimulated or insulin-stimulated TSC2 +/+ or TSC2 −/− MEFs after rapamycin treatment for the indicated times, immunoblotted for phosphotyrosine (panel 1), or the coimmunoprecipitation of the p85 subunit of PI3K (panel 2). The total levels of IRS-1 and p85 in cell lysates are indicated in panels 3 and 4, respectively. (B) Quantitation of insulin- stimulated IRS-1 tyrosine phosphorylation in untreated TSC2 −/− MEFs or the same cells after rapamycin treatment for the indicated times. An arbitrary value was obtained representing the difference between unstimulated and the corresponding insulin-stimulated level of phosphotyrosine in IRS-1 immunoprecipitations as detected on scanned autoradiographs. The values from three independent experiments were averaged and plotted on a bar chart, with the error bars showing the SEM. (C) Phosphorylation of immobilized GST-IRS-1108–516 PTB domain by S6K2 inhibits insulin receptor (InR) interaction. The interaction with tyrosine-phosphorylated InR (top) was determined in vitro by the addition of glutathione beads containing the indicated amounts of S6K2-phosphorylated or unphosphorylated GST-IRS-1108–516 or GST to lysates prepared from insulin-stimulated CHO-IR cells. An immunoblot of the level of GST-IRS-1 fusion protein or GST is indicated on the bottom. DP indicates degradation products of the GST-IRS-1 protein. (D) Quantitation of InR binding to GST-IRS-1108–516 (WT) or a mutant GST-IRS-1108–516 in which serine 302 is mutated to alanine (S302A) after in vitro phosphorylation by S6K2. Error bars show the SEM from three experiments. (E) InR binding of wild-type GST-IRS-1108–516, or S302A and S302E mutants in a pull-down assay. Top, InR; bottom, anti-GST. (F) Quantitation of InR binding to GST-IRS-1108–516 (WT) or S302A and S302E mutants. An arbitrary value of 100 was assigned for the binding of wild-type GST-IRS-1108–516, and the binding of the mutant IRS-1 proteins was expressed relative to this value. Error bars show the SEM from three experiments.
Figure 7.
IGF-1–mediated migration and survival are impaired in TSC2 -deficient MEFs. (A) A chemotaxis assay indicating the migration of TSC2 +/+ or TSC2 −/− MEFs in the absence or presence of 50 ng/ml IGF-1 or 20 ng/ml EGF as a chemotactic factor in a Transwell assay. The pictures show fluorescently labeled cells that have traversed a fluorescently inert filter, such that only the cells that have crossed the filter are photographed. Basal migration (left-hand panels) in the presence of 0.5% FCS present in both chambers and IGF-1–stimulated migration (middle panels) or EGF-stimulated migration (rightmost two panels) is shown in untreated cells (top six panels) or cells pretreated with 20 nM rapamycin for 24 h (bottom four panels). (B) Quantitation of IGF-1– or EGF-mediated chemotaxis in the chemotaxis assay of TSC2 +/+ (gray bars) or TSC2 −/− (white bars) MEFs. The results are expressed as a percentage of that of IGF-1–stimulated chemotaxis of TSC2 +/+ MEFs not treated with rapamycin (100%) from four random 10× fields of two duplicate wells. Error bars show the SEM values. (C) Cell survival assay in the absence of serum. A quantitative analysis of the percentage of TUNEL-positive nuclei is shown for TSC2 +/+ (gray bars) and TSC2 −/− (white bars) MEFs after 8 h in serum-free medium alone (−) or supplemented with 200 ng/ml IGF-1 (+) in untreated MEFs or MEFs pretreated for 36 h with 20 nM rapamycin. The results are from four random 10× fields and are typical of experiments performed twice. (D) Relative cell survival of TSC2 +/+ (gray bars) or TSC2 −/− (white bars) MEFs. The results were obtained by dividing the means of the percentages of TUNEL-positive nuclei in the absence of IGF-1 by the means of those in the presence of IGF-1.
Figure 7.
IGF-1–mediated migration and survival are impaired in TSC2 -deficient MEFs. (A) A chemotaxis assay indicating the migration of TSC2 +/+ or TSC2 −/− MEFs in the absence or presence of 50 ng/ml IGF-1 or 20 ng/ml EGF as a chemotactic factor in a Transwell assay. The pictures show fluorescently labeled cells that have traversed a fluorescently inert filter, such that only the cells that have crossed the filter are photographed. Basal migration (left-hand panels) in the presence of 0.5% FCS present in both chambers and IGF-1–stimulated migration (middle panels) or EGF-stimulated migration (rightmost two panels) is shown in untreated cells (top six panels) or cells pretreated with 20 nM rapamycin for 24 h (bottom four panels). (B) Quantitation of IGF-1– or EGF-mediated chemotaxis in the chemotaxis assay of TSC2 +/+ (gray bars) or TSC2 −/− (white bars) MEFs. The results are expressed as a percentage of that of IGF-1–stimulated chemotaxis of TSC2 +/+ MEFs not treated with rapamycin (100%) from four random 10× fields of two duplicate wells. Error bars show the SEM values. (C) Cell survival assay in the absence of serum. A quantitative analysis of the percentage of TUNEL-positive nuclei is shown for TSC2 +/+ (gray bars) and TSC2 −/− (white bars) MEFs after 8 h in serum-free medium alone (−) or supplemented with 200 ng/ml IGF-1 (+) in untreated MEFs or MEFs pretreated for 36 h with 20 nM rapamycin. The results are from four random 10× fields and are typical of experiments performed twice. (D) Relative cell survival of TSC2 +/+ (gray bars) or TSC2 −/− (white bars) MEFs. The results were obtained by dividing the means of the percentages of TUNEL-positive nuclei in the absence of IGF-1 by the means of those in the presence of IGF-1.
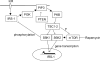
Figure 8.
A model for TSC1-2 regulation of insulin signaling to PI3K. The elevated production of the inositol phospholipid PIP3 by PI3K and its antagonism by PTEN (and 5-phosphatases; not depicted) determines the activation of downstream responses such as activation of PKB. A further downstream response to PKB activation may be the inactivation of TSC1-2 via phosphorylation of tuberin, resulting in S6K activation. In our model, TSC1-2 promotes insulin-like growth factor signaling to PI3K by repressing a negative feedback loop from mTOR/S6K to the adaptor molecule IRS-1. This negative regulation by TSC1-2 may be on mTOR effectors (Jaeschke et al., 2002) or on mTOR directly (Tee et al., 2002). A similar negative feedback involving S6K repression of PKB activation has also been described in Drosophila (Radimerski et al., 2002), although the target of dS6K's inhibitory action here is not known. Repression of IRS-1 by mTOR/S6K occurs via both a transcriptional repression of IRS-1 gene expression mediated by both S6K1 and S6K2 and by direct phosphorylation of IRS-1 protein close to the PTB domain by S6K1. Therefore, failure to activate PI3K may restrict the malignant potential of _TSC_-defective cells.
Similar articles
- Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent.
Jaeschke A, Hartkamp J, Saitoh M, Roworth W, Nobukuni T, Hodges A, Sampson J, Thomas G, Lamb R. Jaeschke A, et al. J Cell Biol. 2002 Oct 28;159(2):217-24. doi: 10.1083/jcb.jcb.200206108. Epub 2002 Oct 28. J Cell Biol. 2002. PMID: 12403809 Free PMC article. - Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies.
Shah OJ, Wang Z, Hunter T. Shah OJ, et al. Curr Biol. 2004 Sep 21;14(18):1650-6. doi: 10.1016/j.cub.2004.08.026. Curr Biol. 2004. PMID: 15380067 - Insulin signaling during perinatal liver development in the rat.
Anand P, Boylan JM, Ou Y, Gruppuso PA. Anand P, et al. Am J Physiol Endocrinol Metab. 2002 Oct;283(4):E844-52. doi: 10.1152/ajpendo.00111.2002. Am J Physiol Endocrinol Metab. 2002. PMID: 12217903 - The mTOR/S6K signalling pathway: the role of the TSC1/2 tumour suppressor complex and the proto-oncogene Rheb.
Nobukini T, Thomas G. Nobukini T, et al. Novartis Found Symp. 2004;262:148-54; discussion 154-9, 265-8. Novartis Found Symp. 2004. PMID: 15562827 Review. - Divergent roles for IRS-1 and IRS-2 in breast cancer metastasis.
Gibson SL, Ma Z, Shaw LM. Gibson SL, et al. Cell Cycle. 2007 Mar 15;6(6):631-7. doi: 10.4161/cc.6.6.3987. Epub 2007 Mar 7. Cell Cycle. 2007. PMID: 17361103 Review.
Cited by
- Inhibition of AMPK catabolic action by GSK3.
Suzuki T, Bridges D, Nakada D, Skiniotis G, Morrison SJ, Lin JD, Saltiel AR, Inoki K. Suzuki T, et al. Mol Cell. 2013 May 9;50(3):407-19. doi: 10.1016/j.molcel.2013.03.022. Epub 2013 Apr 25. Mol Cell. 2013. PMID: 23623684 Free PMC article. - Targeting RAS mutants in malignancies: successes, failures, and reasons for hope.
Yang H, Zhou X, Fu D, Le C, Wang J, Zhou Q, Liu X, Yuan Y, Ding K, Xiao Q. Yang H, et al. Cancer Commun (Lond). 2023 Jan;43(1):42-74. doi: 10.1002/cac2.12377. Epub 2022 Oct 31. Cancer Commun (Lond). 2023. PMID: 36316602 Free PMC article. Review. - The functions and regulation of the PTEN tumour suppressor.
Song MS, Salmena L, Pandolfi PP. Song MS, et al. Nat Rev Mol Cell Biol. 2012 Apr 4;13(5):283-96. doi: 10.1038/nrm3330. Nat Rev Mol Cell Biol. 2012. PMID: 22473468 Review. - The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5'TOP sequence.
Lahr RM, Mack SM, Héroux A, Blagden SP, Bousquet-Antonelli C, Deragon JM, Berman AJ. Lahr RM, et al. Nucleic Acids Res. 2015 Sep 18;43(16):8077-88. doi: 10.1093/nar/gkv748. Epub 2015 Jul 22. Nucleic Acids Res. 2015. PMID: 26206669 Free PMC article. - Capivasertib combines with docetaxel to enhance anti-tumour activity through inhibition of AKT-mediated survival mechanisms in prostate cancer.
Eberlein C, Williamson SC, Hopcroft L, Ros S, Moss JI, Kerr J, van Weerden WM, de Bruin EC, Dunn S, Willis B, Ross SJ, Rooney C, Barry ST. Eberlein C, et al. Br J Cancer. 2024 May;130(8):1377-1387. doi: 10.1038/s41416-024-02614-w. Epub 2024 Feb 23. Br J Cancer. 2024. PMID: 38396173 Free PMC article.
References
- Brown, E.J., P.A. Beal, C.T. Keith, J. Chen, T.B. Shin, and S.L. Schreiber. 1995. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature. 377:441–446. - PubMed
- Cantley, L.C. 2002. The phosphoinositide 3-kinase pathway. Science. 296:1655–1657. - PubMed
- Crackower, M.A., G.Y. Oudit, I. Kozieradzki, R. Sarao, H. Sun, T. Sasaki, E. Hirsch, A. Suzuki, T. Shioi, J. Irie-Sasaki, et al. 2002. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell. 110:737–749. - PubMed
- de Groot, R.P., L.M. Ballou, and P. Sassone-Corsi. 1994. Positive regulation of the cAMP-responsive activator CREM by the p70 S6 kinase: an alternative route to mitogen-induced gene expression. Cell. 79:81–91. - PubMed
- Dufner, A., and G. Thomas. 1999. Ribosomal S6 kinase signaling and the control of translation. Exp. Cell Res. 253:100–109. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases