An amyloid-forming segment of beta2-microglobulin suggests a molecular model for the fibril - PubMed (original) (raw)
An amyloid-forming segment of beta2-microglobulin suggests a molecular model for the fibril
Magdalena I Ivanova et al. Proc Natl Acad Sci U S A. 2004.
Abstract
In humans suffering from dialysis-related amyloidosis, the protein beta2-microglobulin (beta2M) is deposited as an amyloid; however, an amyloid of beta2M is unknown in mice. beta2M sequences from human and mouse are 70% identical, but there is a seven-residue peptide in which six residues differ. This peptide from human beta2M forms amyloid in vitro, whereas the mouse peptide does not. Substitution of the human peptide for its counterpart in the mouse sequence results in the formation of amyloid in vitro. These results show that a seven-residue segment of human beta2M is sufficient to convert beta2M to the amyloid state, and that specific residue interactions are crucial to the conversion. These observations are consistent with a proposed Zipper-spine model for beta2M amyloid, in which the spine of the fibril consists of an anhydrous beta-sheet.
Figures
Fig. 1.
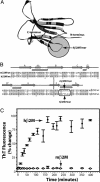
Structure and amyloid formation of hβ2M and mβ2M. (A) In the ribbon diagram of the hβ2M structure (PDB 1LDS), the segments of identical sequence in hβ2M and mβ2M are shown in dark gray. The amyloid-forming hβM7mer (within the shaded circle) is the loop nearest to the C terminus, connecting β-strands F and G. The disulfide bond between Cys 25 and Cys 80 is shown as ball-and-stick. (B) The alignment of the sequences shows that hβ2M and mβ2M are 70% identical. Residues 83–89 (in the oval) form the longest segment, in which the hβ2M and mβ2M sequences differ ( indicates β-strands;
indicates β-strands;  indicates the β-hairpin). (C) The amyloid-forming properties of hβ2M and mβ2M. hβ2M forms amyloid (filled circles), as judged by the characteristic fluorescence after staining the sample with ThT. In contrast, under the same conditions, mβM forms amorphous aggregates (open circles) and does not bind ThT [drawn with
indicates the β-hairpin). (C) The amyloid-forming properties of hβ2M and mβ2M. hβ2M forms amyloid (filled circles), as judged by the characteristic fluorescence after staining the sample with ThT. In contrast, under the same conditions, mβM forms amorphous aggregates (open circles) and does not bind ThT [drawn with
molscript
(35) and
secseq
(
http://xray.imsb.au.dk/\~deb/secseq
)].
Fig. 3.
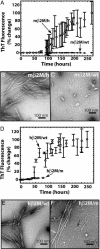
Amyloid fibril formation of wild-type and chimeric β2M. (A) mβ2M/wt (filled circles) does not bind ThT. In contrast, the mβ2M/h aggregates (open circles) bind ThT starting ≈4 days after initiating the reaction. This demonstrates the ability of the human heptamer to promote mβ2M to form amyloid-like aggregates. (B) The mβ2M/h sample consists of long straight fibrils typical for amyloids. (C) No fibrils were observed in the mβ2M/wt sample, and the largest aggregates observed appear globular, as indicated by the white arrows. Micrographs shown in B and C were taken 14 days after the initiation of the aggregation reaction. (D) Fibril formation of hβ2M is disrupted when the mβ2M7mer is swapped into the hβ2M scaffold. Aggregation of the hβ2M/m chimera (open circles) lags behind that of the hβ2M/wt (filled circles) fibril formation. hβ2M/wt reaches the aggregated state ≈3–24 h earlier than hβ2M/m. The fluorescence of the two proteins after the aggregated state is reached is indistinguishable. (E) hβ2M/wt sample had a homogenous population of long and straight fibrils, typical for amyloid, and no amorphous aggregates were observed in the sample. (F) In contrast, hβ2M/m sample displayed equal populations of long (white arrow) and short fibrils (black arrow). The electron micrographs of samples shown in E and F were taken 10 days after the initiation of the aggregation reaction.
Fig. 2.
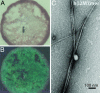
Amyloid fiber formation by hβ2M7mer. (A) hβ2M7mer fibrils stained with Congo red solution appear red when viewed under unpolarized light. (B) When viewed between crosspolarizers, the sample shown in A exhibits “apple green” birefringence typical for amyloid fibrils. (C) Samples of hβM7mer contain fibrils when viewed by EM.
Fig. 4.
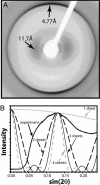
X-ray diffraction cross β pattern of β2M/wt fibrils. (A) The characteristic reflections for amyloid fibrils are indicated by arrows. (B) Comparison of the observed profile of the ≈11-Å diffuse reflection with profiles calculated from models representing the Zipper-spine as one to four slits (β-sheets).
Fig. 5.
Speculative Zipper-spine model for the β2M protofilament. (A) The C-terminal segment of β2M is the portion of the structure to rearrange during fibril formation and the amyloid-forming hβ2M7mer (boxed) forms a new β-strand F′.(B) In the fibril model, residues Pro 90 and Lys 91 form a type I β-turn. Thus the β-strand G is hydrogen bonded with the new β-strand F′ rather than with the β-strand F of the native structure. (C) Two such F′-G hairpins stack to form the asymmetric unit of one of the sheets of the spine. Each sheet is built with a small (7°) twist between β-strands, stacked 4.7 Å apart, so that the spine has a pitch of ≈242 Å. We expect these parameters will be refined as structural data become available. The sheets are separated by ≈11 Å. β-Strands A–E and part of F retain their native structure with the disulfide bond between β-strands B and F intact. These native-like core domains decorate the periphery of the double β-sheet spine, with only four molecules shown for clarity. (D) The view down the fiber axis shows the double β-sheet spine. The rest of the β2M molecules, which remain folded, are packed closely around the β-sheet spine. (E) A space-filling model (view of the fibril down its axis) shows that each of the four molecules, represented with different colors, is tightly packed within the fibril, with no space for solvent. Thus, the core domains shield the spine from solvent.
Similar articles
- Structure of interacting segments in the growing amyloid fibril of beta2-microglobulin probed with IR spectroscopy.
Lu M, Hiramatsu H, Goto Y, Kitagawa T. Lu M, et al. J Mol Biol. 2006 Sep 15;362(2):355-64. doi: 10.1016/j.jmb.2006.07.023. Epub 2006 Jul 15. J Mol Biol. 2006. PMID: 16919295 - Role of the C-terminal 28 residues of beta2-microglobulin in amyloid fibril formation.
Ivanova MI, Gingery M, Whitson LJ, Eisenberg D. Ivanova MI, et al. Biochemistry. 2003 Nov 25;42(46):13536-40. doi: 10.1021/bi0301486. Biochemistry. 2003. PMID: 14622000 - Seeding-dependent propagation and maturation of amyloid fibril conformation.
Yamaguchi K, Takahashi S, Kawai T, Naiki H, Goto Y. Yamaguchi K, et al. J Mol Biol. 2005 Sep 30;352(4):952-60. doi: 10.1016/j.jmb.2005.07.061. J Mol Biol. 2005. PMID: 16126222 - Molecular interactions in the formation and deposition of beta2-microglobulin-related amyloid fibrils.
Naiki H, Yamamoto S, Hasegawa K, Yamaguchi I, Goto Y, Gejyo F. Naiki H, et al. Amyloid. 2005 Mar;12(1):15-25. doi: 10.1080/13506120500032352. Amyloid. 2005. PMID: 16076607 Review. - Limited proteolysis in the investigation of beta2-microglobulin amyloidogenic and fibrillar states.
Monti M, Amoresano A, Giorgetti S, Bellotti V, Pucci P. Monti M, et al. Biochim Biophys Acta. 2005 Nov 10;1753(1):44-50. doi: 10.1016/j.bbapap.2005.09.004. Epub 2005 Sep 23. Biochim Biophys Acta. 2005. PMID: 16213198 Review.
Cited by
- Critical structural and functional roles for the N-terminal insertion sequence in surfactant protein B analogs.
Walther FJ, Waring AJ, Hernandez-Juviel JM, Gordon LM, Wang Z, Jung CL, Ruchala P, Clark AP, Smith WM, Sharma S, Notter RH. Walther FJ, et al. PLoS One. 2010 Jan 13;5(1):e8672. doi: 10.1371/journal.pone.0008672. PLoS One. 2010. PMID: 20084172 Free PMC article. - Distinguishing closely related amyloid precursors using an RNA aptamer.
Sarell CJ, Karamanos TK, White SJ, Bunka DHJ, Kalverda AP, Thompson GS, Barker AM, Stockley PG, Radford SE. Sarell CJ, et al. J Biol Chem. 2014 Sep 26;289(39):26859-26871. doi: 10.1074/jbc.M114.595066. Epub 2014 Aug 6. J Biol Chem. 2014. PMID: 25100729 Free PMC article. - CORDAX web server: an online platform for the prediction and 3D visualization of aggregation motifs in protein sequences.
Louros N, Rousseau F, Schymkowitz J. Louros N, et al. Bioinformatics. 2024 May 2;40(5):btae279. doi: 10.1093/bioinformatics/btae279. Bioinformatics. 2024. PMID: 38662570 Free PMC article. - Staphylococcal Bap Proteins Build Amyloid Scaffold Biofilm Matrices in Response to Environmental Signals.
Taglialegna A, Navarro S, Ventura S, Garnett JA, Matthews S, Penades JR, Lasa I, Valle J. Taglialegna A, et al. PLoS Pathog. 2016 Jun 21;12(6):e1005711. doi: 10.1371/journal.ppat.1005711. eCollection 2016 Jun. PLoS Pathog. 2016. PMID: 27327765 Free PMC article. - Decoding the Structural Bases of D76N ß2-Microglobulin High Amyloidogenicity through Crystallography and Asn-Scan Mutagenesis.
de Rosa M, Barbiroli A, Giorgetti S, Mangione PP, Bolognesi M, Ricagno S. de Rosa M, et al. PLoS One. 2015 Dec 1;10(12):e0144061. doi: 10.1371/journal.pone.0144061. eCollection 2015. PLoS One. 2015. PMID: 26625273 Free PMC article.
References
- Rudall, K. M. (1952) Adv. Protein Chem. 7, 253–290. - PubMed
- Westermark, P., Benson, M. D., Buxbaum, J. N., Cohen, A. S., Frangione, B., Ikeda, S., Masters, C. L., Merlini, G., Saraiva, M. J. & Sipe, J. D. (2002) Amyloid 9, 197–200. - PubMed
- Dobson, C. M. (1999) Trends Biochem. Sci. 24, 329–332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources