Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif - PubMed (original) (raw)
Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif
Björn Schwer et al. J Virol. 2004 Aug.
Abstract
The hepatitis C virus (HCV) core protein represents the first 191 amino acids of the viral precursor polyprotein and is cotranslationally inserted into the membrane of the endoplasmic reticulum (ER). Processing at position 179 by a recently identified intramembrane signal peptide peptidase leads to the generation and potential cytosolic release of a 179-amino-acid matured form of the core protein. Using confocal microscopy, we observed that a fraction of the mature core protein colocalized with mitochondrial markers in core-expressing HeLa cells and in Huh-7 cells containing the full-length HCV replicon. Subcellular fractionation confirmed this observation and showed that the core protein associates with purified mitochondrial fractions devoid of ER contaminants. The core protein also fractionated with mitochondrion-associated membranes, a site of physical contact between the ER and mitochondria. Using immunoelectron microscopy and in vitro mitochondrial import assays, we showed that the core protein is located on the mitochondrial outer membrane. A stretch of 10 amino acids within the hydrophobic C terminus of the processed core protein conferred mitochondrial localization when it was fused to green fluorescent protein. The location of the core protein in the outer mitochondrial membrane suggests that it could modulate apoptosis or lipid transfer, both of which are associated with this subcellular compartment, during HCV infection.
Figures
FIG. 1.
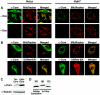
A fraction of the HCV core protein colocalizes with a mitochondrial marker in transfected cells. Confocal microscopic images of HeLa Tet-Off and Huh-7 cells expressing the core protein from a tetracycline-controlled (A) or a CMV-driven expression vector (B) are shown. Living cells were incubated with MitoTracker Red or were cotransfected with a construct expressing the ER-targeted Discosoma red fluorescent protein (DsRed-ER). Immunostaining for the core protein was performed with anti-core protein and Cy2-labeled anti-mouse antibodies 20 h after transfection. (C) Western blot analysis of core protein expressed from the tetracycline-controlled or CMV-driven vector. The blot was stripped and reprobed for tubulin as a loading control. (D) Comparison of core protein expressed from the CMV-driven vector with in vitro-synthesized truncated core protein (173 amino acids) and unprocessed core protein (191 amino acids) generated in the absence of microsomal membranes.
FIG. 2.
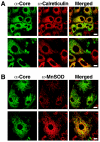
Partial localization of core protein to mitochondria in Huh-7 cells expressing a full-length HCV replicon. Huh-7 cells (clone 21-5) containing a full-length HCV replicon were costained for the core protein and calreticulin (A) or the core protein and MnSOD (B). Bars: 20 μm in upper panel and 10 μm in lower panel (A) or 20 μm (B).
FIG. 3.
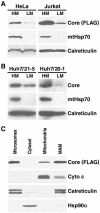
Core protein cofractionates with mitochondria. (A) Western blot analysis of HM and LM fractions isolated from core protein-expressing HeLa and Jurkat Tet-Off cells by differential centrifugation. Cell equivalents from each fraction were loaded, and the distributions of FLAG-tagged core protein, mitochondrial heat shock protein 70 (mtHsp70), and the ER-resident calreticulin protein were analyzed. (B) Western blot analysis of HM and LM fractions of two Huh-7 cell lines (21-5 and 20-1) expressing the full-length replicon with core protein- and organelle-specific antibodies. (C) Jurkat homogenates were further separated by sucrose density gradient centrifugation to obtain purified mitochondria and MAMs. Microsomes and cytosolic fractions were prepared by differential centrifugation of the same cell homogenates. Equal protein amounts (30 μg) from each fraction were analyzed by Western blotting with antibodies specific for FLAG-tagged core protein, cytochrome c (Cyto c), calreticulin, and heat shock protein 90α (Hsp90α).
FIG. 4.
Core protein localizes to the outer surfaces of mitochondria. The images shown are confocal (A) and immunoelectron (B to D) microscopic images of the core protein in HeLa cells transduced with a full-length core protein-expressing lentiviral vector and an immunoelectron microscopic image (E) of the core protein in Huh-7 cells containing a full-length HCV replicon. Core protein-specific gold particles (5-nm diameter) were predominantly found at the outer surfaces of mitochondria (M). For panel E, nanogold particles (1.4-nm diameter) were used and enhanced with a silver enhancement step. Bars: 0.2 μm. IF, intermediate filaments.
FIG. 5.
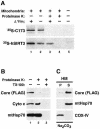
Nonintegral attachment of core protein to the mitochondrial outer membrane. (A) In vitro import assays using isolated mitochondria and in vitro-synthesized radiolabeled core protein (173 amino acids) or human SIRT3 (hSIRT3) were analyzed by autoradiography. The core protein was not imported into mitochondria and was accessible to the proteolytic activity of proteinase K (top, lane 3). SIRT3 was partially protected from digestion by proteinase K because of active mitochondrial import (bottom, lane 3). Accordingly, inhibitors of the mitochondrial membrane potential (Δψm) reduced the signal for SIRT3 (bottom, lane 2) but had no effect on the core protein (top, lane 2). SIRT3 was completely digested by proteinase K when active mitochondrial import was inhibited (bottom, lane 4). (B) Proteinase K digestion of mitochondria isolated from core protein-expressing Jurkat Tet-Off cells in the presence or absence of detergent (TX-100). A Western blot analysis showed that while the FLAG-tagged core protein was readily digested by proteinase K in the absence of TX-100 (lane 2), proteins located in the intermembrane space, such as cytochrome c (Cyto c), and mtHsp70, which is localized in the mitochondrial matrix, were protected. (C) Sodium carbonate extracts of HM fractions isolated from core protein-expressing Jurkat Tet-Off cells were analyzed for the distribution of the FLAG-tagged core protein, mtHsp70, which in the matrix is peripherally attached to the inner mitochondrial membrane, and COX-IV, a polytopic inner mitochondrial membrane protein.
FIG. 6.
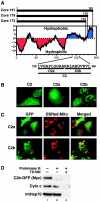
A 10-amino-acid motif functions as a mitochondrial targeting signal in the core protein. (A) Kyte-Doolittle hydrophobicity plot of full-length (Core 191) and processed (Core 179 and Core 173) core proteins. The amino acid sequences and positions of domain C2 and the subdivisions C2a and C2b within the full-length core protein are indicated at the bottom. (B) Confocal microscopy of Huh-7 cells transfected with fusion proteins of domains C2, C2a, or C2b to GFP. (C) Confocal microscopy of Huh-7 cells cotransfected with the indicated GFP fusion proteins and a construct expressing the mitochondrion-targeted Discosoma red fluorescent protein (DsRed-Mito). (D) Proteinase K digestion of crude mitochondrial fractions isolated from Huh-7 cells transfected with C2b-GFP fusion protein in the absence or presence of detergent (TX-100). A Western blot analysis was performed with antibodies against the Myc epitope to detect the Myc-tagged fusion protein, against cytochrome c (Cyto c), and against mtHsp70.
FIG. 7.
Model of mitochondrial translocation of core protein. See the text for details.
Similar articles
- Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein.
Okamoto K, Moriishi K, Miyamura T, Matsuura Y. Okamoto K, et al. J Virol. 2004 Jun;78(12):6370-80. doi: 10.1128/JVI.78.12.6370-6380.2004. J Virol. 2004. PMID: 15163730 Free PMC article. - Molecular determinants for subcellular localization of hepatitis C virus core protein.
Suzuki R, Sakamoto S, Tsutsumi T, Rikimaru A, Tanaka K, Shimoike T, Moriishi K, Iwasaki T, Mizumoto K, Matsuura Y, Miyamura T, Suzuki T. Suzuki R, et al. J Virol. 2005 Jan;79(2):1271-81. doi: 10.1128/JVI.79.2.1271-1281.2005. J Virol. 2005. PMID: 15613354 Free PMC article. - Internally located signal peptides direct hepatitis C virus polyprotein processing in the ER membrane.
Wu JZ. Wu JZ. IUBMB Life. 2001 Jan;51(1):19-23. doi: 10.1080/15216540119497. IUBMB Life. 2001. PMID: 11419691 - Novel targeting signals mediate the sorting of different isoforms of the tail-anchored membrane protein cytochrome b5 to either endoplasmic reticulum or mitochondria.
Hwang YT, Pelitire SM, Henderson MP, Andrews DW, Dyer JM, Mullen RT. Hwang YT, et al. Plant Cell. 2004 Nov;16(11):3002-19. doi: 10.1105/tpc.104.026039. Epub 2004 Oct 14. Plant Cell. 2004. PMID: 15486098 Free PMC article. - Molecular basis of subcellular localization of HCV core protein.
Suzuki T, Matsuura Y, Harada T, Suzuki R, Saito I, Miyamura T. Suzuki T, et al. Liver. 1996 Aug;16(4):221-4. doi: 10.1111/j.1600-0676.1996.tb00732.x. Liver. 1996. PMID: 8877990 Review. No abstract available.
Cited by
- Mitochondrial and secretory human cytomegalovirus UL37 proteins traffic into mitochondrion-associated membranes of human cells.
Bozidis P, Williamson CD, Colberg-Poley AM. Bozidis P, et al. J Virol. 2008 Mar;82(6):2715-26. doi: 10.1128/JVI.02456-07. Epub 2008 Jan 16. J Virol. 2008. PMID: 18199645 Free PMC article. - Enterovirus 71 induces mitochondrial reactive oxygen species generation that is required for efficient replication.
Cheng ML, Weng SF, Kuo CH, Ho HY. Cheng ML, et al. PLoS One. 2014 Nov 17;9(11):e113234. doi: 10.1371/journal.pone.0113234. eCollection 2014. PLoS One. 2014. PMID: 25401329 Free PMC article. - Visualization and analysis of hepatitis C virus structural proteins at lipid droplets by super-resolution microscopy.
Eggert D, Rösch K, Reimer R, Herker E. Eggert D, et al. PLoS One. 2014 Jul 11;9(7):e102511. doi: 10.1371/journal.pone.0102511. eCollection 2014. PLoS One. 2014. PMID: 25019511 Free PMC article. - Viral product trafficking to mitochondria, mechanisms and roles in pathogenesis.
Williamson CD, DeBiasi RL, Colberg-Poley AM. Williamson CD, et al. Infect Disord Drug Targets. 2012 Feb;12(1):18-37. doi: 10.2174/187152612798994948. Infect Disord Drug Targets. 2012. PMID: 22034933 Free PMC article. Review. - EV71 virus reduces Nrf2 activation to promote production of reactive oxygen species in infected cells.
Bai Z, Zhao X, Li C, Sheng C, Li H. Bai Z, et al. Gut Pathog. 2020 Apr 23;12:22. doi: 10.1186/s13099-020-00361-w. eCollection 2020. Gut Pathog. 2020. PMID: 32346399 Free PMC article. Retracted.
References
- Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. - PubMed
- Ardail, D., F. Gasnier, F. Lerme, C. Simonot, P. Louisot, and O. Gateau-Roesch. 1993. Involvement of mitochondrial contact sites in the subcellular compartmentalization of phospholipid biosynthetic enzymes. J. Biol. Chem. 268:25985-25992. - PubMed
- Ardail, D., F. Lerme, and P. Louisot. 1991. Involvement of contact sites in phosphatidylserine import into liver mitochondria. J. Biol. Chem. 266:7978-7981. - PubMed
- Barba, G., F. Harper, T. Harada, M. Kohara, S. Goulinet, Y. Matsuura, G. Eder, Z. Schaff, M. J. Chapman, T. Miyamura, and C. Brechot. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates with cellular lipid storage droplets. Proc. Natl. Acad. Sci. USA 94:1200-1205. - PMC - PubMed
- Barbaro, G., G. Di Lorenzo, A. Asti, M. Ribersani, G. Belloni, B. Grisorio, G. Filice, and G. Barbarini. 1999. Hepatocellular mitochondrial alterations in patients with chronic hepatitis C: ultrastructural and biochemical findings. Am. J. Gastroenterol. 94:2198-2205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous