Rap1 regulates the formation of E-cadherin-based cell-cell contacts - PubMed (original) (raw)
Rap1 regulates the formation of E-cadherin-based cell-cell contacts
Catherine Hogan et al. Mol Cell Biol. 2004 Aug.
Abstract
In epithelial tissues, cells are linked to their neighbors through specialized cell-cell adhesion proteins. E-cadherin is one of the most important membrane proteins for the establishment of intimate cell-cell contacts, but the molecular mechanism by which it is recruited to contact sites is largely unknown. We report here that the cytoplasmic domain of E-cadherin interacts with C3G, a guanine nucleotide exchange factor for Rap1. In epithelial cell cultures, ligation of the extracellular domain of E-cadherin enhances Rap1 activity, which in turn is necessary for the proper targeting of E-cadherin molecules to maturing cell-cell contacts. Furthermore, our data suggest that Cdc42 functions downstream of Rap1 in this process. We conclude that Rap1 plays a vital role in the establishment of E-cadherin-based cell-cell adhesion.
Figures
FIG. 1.
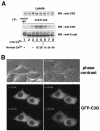
Interaction between C3G and E-cadherin. (A) Molecular domain structure of C3G. The clone isolated by the yeast two-hybrid screen encodes the N terminus of C3G (amino acids 144 to 230). (B) Summary of interactions between cadherins and C3G in yeast two-hybrid assays. The CH of CH2 and CH3 domains stands for cadherin homology (32). The CH2 and CH3 domains are conserved between classical cadherins and bind to p120ctn and β-catenin, respectively. (C) Interaction between C3G and E-cadherin by GST pulldown assay. Flag-tagged C3G (amino acids 1 to 357) expressed in HEK293 cells was pulled down by GST- or GST-E-cadherin (cytoplasmic domain)-coated beads. (D) Direct interaction between C3G and E-cadherin by overlay-blotting assay. Flag-tagged C3G (full length) expressed in HEK293 cells was immunoprecipitated, followed by SDS-PAGE and overlay-blotting assay with the purified recombinant E-cadherin protein (left panel). E-cadherin bound to C3G on the membrane was detected by anti-E-cadherin antibody. The same immunoprecipitated samples were also examined by Western blotting with anti-Flag antibody (right panel). The arrowhead indicates the position of immunoglobulin G heavy chains from anti-Flag antibody. (E) Specificity of the association between E-cadherin and other GEFs. E-cadherin and HA-PDZ-GEF or HA-Rlf were coexpressed in HEK293 cells, and the interaction was examined by immunoprecipitation with anti-E-cadherin antibody. (F) Competition between C3G and β-catenin for binding to E-cadherin. β-catenin or p120ctn was coexpressed with E-cadherin and Flag-C3G (amino acids 1 to 357), and the levels of C3G present in E-cadherin immunoprecipitates were examined.
FIG. 1.
Interaction between C3G and E-cadherin. (A) Molecular domain structure of C3G. The clone isolated by the yeast two-hybrid screen encodes the N terminus of C3G (amino acids 144 to 230). (B) Summary of interactions between cadherins and C3G in yeast two-hybrid assays. The CH of CH2 and CH3 domains stands for cadherin homology (32). The CH2 and CH3 domains are conserved between classical cadherins and bind to p120ctn and β-catenin, respectively. (C) Interaction between C3G and E-cadherin by GST pulldown assay. Flag-tagged C3G (amino acids 1 to 357) expressed in HEK293 cells was pulled down by GST- or GST-E-cadherin (cytoplasmic domain)-coated beads. (D) Direct interaction between C3G and E-cadherin by overlay-blotting assay. Flag-tagged C3G (full length) expressed in HEK293 cells was immunoprecipitated, followed by SDS-PAGE and overlay-blotting assay with the purified recombinant E-cadherin protein (left panel). E-cadherin bound to C3G on the membrane was detected by anti-E-cadherin antibody. The same immunoprecipitated samples were also examined by Western blotting with anti-Flag antibody (right panel). The arrowhead indicates the position of immunoglobulin G heavy chains from anti-Flag antibody. (E) Specificity of the association between E-cadherin and other GEFs. E-cadherin and HA-PDZ-GEF or HA-Rlf were coexpressed in HEK293 cells, and the interaction was examined by immunoprecipitation with anti-E-cadherin antibody. (F) Competition between C3G and β-catenin for binding to E-cadherin. β-catenin or p120ctn was coexpressed with E-cadherin and Flag-C3G (amino acids 1 to 357), and the levels of C3G present in E-cadherin immunoprecipitates were examined.
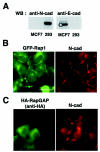
FIG. 2.
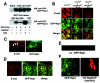
Interaction and localization of C3G and E-cadherin in MCF7 cells. (A) Interaction between endogenous C3G and E-cadherin. MCF7 cells were incubated in low-calcium medium and then transferred to normal-calcium medium for the indicated times. Coimmunoprecipitation of endogenous C3G with E-cadherin was examined. Lanes: 2, no calcium switch; 1 and 3, low calcium; 4 to 8, switch from low calcium to normal calcium (lane 4, 15 min; lane 5, 30 min; lane 6, 1 h; lane 7, 2 h; lane 8, 4 h). (B) Localization of C3G during induction of cell-cell contacts. GFP-tagged C3G (full length) was transiently expressed in MCF7 cells, and the localization of C3G during calcium switch was analyzed by time-lapse images. Upper panel, phase contrast images. Lower panels, GFP-C3G. Arrows indicate the positions of transfected cells.
FIG. 3.
Recruitment of Rap1 at cell-cell contact sites. (A) Characterization of GFP-Rap1 (wild type). GFP-Rap1 was coexpressed with RapGAP or C3G, and active GTP-bound GFP-Rap1 was pulled down by GST-RalGDS beads. (B and D) Colocalization of Rap1 with E-cadherin. MCF7 cells stably expressing GFP-Rap1 were incubated in a low-calcium medium and transferred to a normal-calcium medium for the indicated time, and colocalization of Rap1 and E-cadherin was examined by epifluorescent microscopy (B; 30 min and 2 h) or by confocal microscopy (D; 2 h). (C) Magnified images of GFP-Rap1 and E-cadherin from Fig. 4B (middle top and middle center panels). Arrowheads indicate zipper-like structures of newly formed cell-cell contact sites. (E) Vertical section (XZ) analysis of the localization of GFP-Rap1 in MCF7 cells. (F) Effect of overexpression of RapGAP on localization of GFP-Rap1 after the calcium switch. RapGAP cDNA was microinjected into MCF7 cells stably expressing GFP-Rap1, and the localization of Rap1 was examined 30 min after calcium restoration. Bars, 30 μm.
FIG. 4.
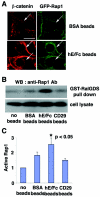
Effect of the establishment of E-cadherin-based cell-cell contacts on the localization and activation of Rap1. (A) Recruitment of Rap1 at E-cadherin-based cell-cell contact sites induced by the extracellular domain of E-cadherin (hE/Fc)-coated beads. BSA- or hE/Fc-coated beads were plated on MCF7 cells expressing GFP-Rap1. After 10 min, the recruitment of β-catenin or GFP-Rap1 to cell-bead contact sites was examined by confocal microscopy. The arrows indicate the positions of the beads. Bar, 30 μm. (B) Activation of Rap1 induced by hE/Fc beads. BSA-, hE/Fc-, or anti-integrin-β1 (CD29) antibody-coated beads were plated on MCF7 cells. Under the experimental conditions, approximately 70% of the cells were in contact with the beads. After 10 min, the cell lysates were used in a GST-RalGDS pulldown assay, followed by Western blotting with anti-Rap1 antibody. (C) Quantitative data for activation of Rap1 by the beads. The relative amount of active Rap1 in each sample was quantified by densitometry and corrected for the total Rap1 in the cell lysate. Results are expressed relative to the no-beads control value and represent the means ± standard deviation of five independent experimental determinations. *, significantly higher than the control (P < 0.05).
FIG. 5.
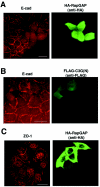
Role of Rap1 in E-cadherin localization. After microinjection, MCF7 cells were subjected to a calcium switch, and the effect of expression of different molecules on the localization of E-cadherin (A and B, 30 min after calcium restoration) or ZO-1 (C, 60 min after calcium restoration) was examined by immunofluorescence. The following cDNAs were microinjected: (A and C) RapGAP; (B) C3G(N) (amino acids 1 to 357). Bars, 30 μm.
FIG. 6.
Role of Rap and Cdc42 in E-cadherin localization. (A and B) Constitutively active Cdc42 enhanced the recruitment of E-cadherin into newly formed cell-cell contacts. After microinjection, MCF7 cells were subjected to a calcium switch, and the effect of expression of different molecules on the localization of E-cadherin was examined 30 min after calcium restoration by immunofluorescence. The following cDNAs were microinjected: (A) constitutively active Cdc42 (Cdc42L61); (B) Cdc42L61 plus RapGAP. Bar, 30 μm. (C) Expression of RapGAP inhibited the activation of Cdc42 during calcium switch. Wild-type Cdc42 (Myc tagged) was transiently expressed with or without RapGAP in MCF7 cells. After the calcium switch, the cell lysates were used in a GST-WASP-CRIB pulldown assay, followed by Western blotting with anti-Myc antibody. The time indicates the time after the calcium switch. (D) Quantitative data for activation of Cdc42 during the calcium switch. The relative amount of active Myc-Cdc42 in each sample was quantified by densitometry and corrected for the total Myc-Cdc42 in the cell lysate. Results are expressed relative to the time zero value and represent the means ± standard deviation of several independent experimental determinations (n = 7 for 0, 30, and 60 min; n = 4 for 120 min). * and **, significantly higher without RapGAP than with RapGAP (*, P < 0.002; **, P < 0.0005).
FIG. 7.
Localization and role of Rap1 in N-cadherin-based cell-cell contacts in HEK293 cells. (A) N-cadherin but not E-cadherin is expressed in HEK293 cells. Cell lysates from HEK293 and MCF7 cells were examined for expression of E-cadherin and N-cadherin by Western blotting. (B) GFP-Rap1 is colocalized with N-cadherin at cell-cell contact sites in HEK293 cells after a calcium switch. GFP-Rap1 was transiently expressed in HEK293 cells, and the localization of GFP-Rap1 and N-cadherin was examined 1 h after calcium restoration. (C) The expression of RapGAP does not affect the localization of N-cadherin at cell-cell contact sites after a calcium switch. RapGAP was transiently expressed in HEK293 cells, and the localization of N-cadherin was examined 1 h after calcium restoration. Bar, 30 μm.
Similar articles
- Cyclic AMP potentiates vascular endothelial cadherin-mediated cell-cell contact to enhance endothelial barrier function through an Epac-Rap1 signaling pathway.
Fukuhara S, Sakurai A, Sano H, Yamagishi A, Somekawa S, Takakura N, Saito Y, Kangawa K, Mochizuki N. Fukuhara S, et al. Mol Cell Biol. 2005 Jan;25(1):136-46. doi: 10.1128/MCB.25.1.136-146.2005. Mol Cell Biol. 2005. PMID: 15601837 Free PMC article. - Activation of Rac by cadherin through the c-Src-Rap1-phosphatidylinositol 3-kinase-Vav2 pathway.
Fukuyama T, Ogita H, Kawakatsu T, Inagaki M, Takai Y. Fukuyama T, et al. Oncogene. 2006 Jan 5;25(1):8-19. doi: 10.1038/sj.onc.1209010. Oncogene. 2006. PMID: 16170364 - E-cadherin dis-engagement activates the Rap1 GTPase.
Asuri S, Yan J, Paranavitana NC, Quilliam LA. Asuri S, et al. J Cell Biochem. 2008 Nov 1;105(4):1027-37. doi: 10.1002/jcb.21902. J Cell Biochem. 2008. PMID: 18767072 Free PMC article. - Vascular endothelial cadherin-mediated cell-cell adhesion regulated by a small GTPase, Rap1.
Fukuhra S, Sakurai A, Yamagishi A, Sako K, Mochizuki N. Fukuhra S, et al. J Biochem Mol Biol. 2006 Mar 31;39(2):132-9. doi: 10.5483/bmbrep.2006.39.2.132. J Biochem Mol Biol. 2006. PMID: 16584626 Review. - Rap1: a key regulator in cell-cell junction formation.
Kooistra MR, Dubé N, Bos JL. Kooistra MR, et al. J Cell Sci. 2007 Jan 1;120(Pt 1):17-22. doi: 10.1242/jcs.03306. J Cell Sci. 2007. PMID: 17182900 Review.
Cited by
- Syndecan-1 controls cell migration by activating Rap1 to regulate focal adhesion disassembly.
Altemeier WA, Schlesinger SY, Buell CA, Parks WC, Chen P. Altemeier WA, et al. J Cell Sci. 2012 Nov 1;125(Pt 21):5188-95. doi: 10.1242/jcs.109884. Epub 2012 Aug 16. J Cell Sci. 2012. PMID: 22899717 Free PMC article. - Discontinuities in Rap1 activity determine epithelial cell morphology within the developing wing of Drosophila.
O'Keefe DD, Gonzalez-Niño E, Edgar BA, Curtiss J. O'Keefe DD, et al. Dev Biol. 2012 Sep 15;369(2):223-34. doi: 10.1016/j.ydbio.2012.06.024. Epub 2012 Jul 7. Dev Biol. 2012. PMID: 22776378 Free PMC article. - Cyclic AMP potentiates vascular endothelial cadherin-mediated cell-cell contact to enhance endothelial barrier function through an Epac-Rap1 signaling pathway.
Fukuhara S, Sakurai A, Sano H, Yamagishi A, Somekawa S, Takakura N, Saito Y, Kangawa K, Mochizuki N. Fukuhara S, et al. Mol Cell Biol. 2005 Jan;25(1):136-46. doi: 10.1128/MCB.25.1.136-146.2005. Mol Cell Biol. 2005. PMID: 15601837 Free PMC article. - Integrin Regulators in Neutrophils.
Pulikkot S, Hu L, Chen Y, Sun H, Fan Z. Pulikkot S, et al. Cells. 2022 Jun 25;11(13):2025. doi: 10.3390/cells11132025. Cells. 2022. PMID: 35805108 Free PMC article. Review. - Epac-Rap signaling reduces cellular stress and ischemia-induced kidney failure.
Stokman G, Qin Y, Genieser HG, Schwede F, de Heer E, Bos JL, Bajema IM, van de Water B, Price LS. Stokman G, et al. J Am Soc Nephrol. 2011 May;22(5):859-72. doi: 10.1681/ASN.2010040423. Epub 2011 Apr 14. J Am Soc Nephrol. 2011. PMID: 21493776 Free PMC article.
References
- Adams, C. L., and W. J. Nelson. 1998. Cytomechanics of cadherin-mediated cell-cell adhesion. Curr. Opin. Cell Biol. 10:572-577. - PubMed
- Bos, J. L., J. de Rooij, and K. A. Reedquist. 2001. Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell. Biol. 2:369-377. - PubMed
- Braga, V. M. M. 2002. Cadherin adhesion regulation in keratinocytes, p. 1-36. In T. P. Fleming (ed.), Cell-cell interactions: a practical approach, 2nd ed. Oxford University Press, Oxford, England.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous