Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line - PubMed (original) (raw)
Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line
Alexei A Aravin et al. Mol Cell Biol. 2004 Aug.
Abstract
To date, few natural cases of RNA-silencing-mediated regulation have been described. Here, we analyzed repression of testis-expressed Stellate genes by the homologous Suppressors of Stellate [Su(Ste)] repeats that produce sense and antisense short RNAs. The Stellate promoter is dispensable for suppression, but local disturbance of complementarity between the Stellate transcript and the Su(Ste) repeats impairs silencing. Using in situ RNA hybridization, we found temporal control of the expression and spatial distribution of sense and antisense Stellate and Su(Ste) transcripts in germinal cells. Antisense Su(Ste) transcripts accumulate in the nuclei of early spermatocytes before the appearance of sense transcripts. The sense and antisense transcripts are colocalized in the nuclei of mature spermatocytes, placing the initial step of silencing in the nucleus and suggesting formation of double-stranded RNA. Mutations in the aubergine and spindle-E genes, members of the Argonaute and RNA helicase gene families, respectively, impair silencing by eliminating the short Su(Ste) RNA, but have no effect on microRNA production. Thus, different small RNA-containing complexes operate in the male germ line.
Figures
FIG. 1.
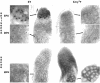
Localization of sense and antisense Stellate and Su(Ste) transcripts in testes. Sense and antisense RNA were detected by using single-stranded probes in whole testes of wild-type males (XY) and of males with a deletion of the bulk of Su(Ste) repeats (Xcry1Y). DIG-labeled probes were visualized by using AP-coupled antibodies following color reaction with NBT/BCIP substrate. The segments shown in enlarged panels correspond to early (EPS) and mature (MPS) primary spermatocyte stages. In wild-type males, antisense transcripts are abundant in the nuclei of early primary spermatocytes, while in cry1Y males the signal area is greatly decreased. In mature primary spermatocytes, antisense transcripts are detected as one or two sharp dots per nucleus. In cry1Y males the signals are not visible in all nuclei. Sense RNA is detected only at the mature primary spermatocyte stage, as faint dots in nuclei of the wild-type males or as a strong diffuse signal in the cytoplasm of cry1Y males.
FIG. 2.
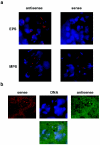
FISH detection of sense and antisense transcripts in wild-type spermatocytes. (a) Sense and antisense RNAs in the nuclei of early (EPS) and mature (MPS) primary spermatocytes of wild-type males. DIG-labeled probes were visualized by using rhodamine-coupled antibodies (red); DNA was stained with DAPI (blue). Staining in the cytoplasm is due to tissue autofluorescence. (b) Simultaneous detection of sense and antisense transcripts in mature primary spermatocyte nuclei. The DIG-labeled probe for sense RNA was visualized with rhodamine-coupled antibodies (red), and the biotin-labeled probe for antisense RNA was visualized with fluorescein isothiocyanate-coupled antibodies (green). DNA was stained with DAPI (blue). Images obtained from separate channels (upper line) and the composite image (bottom) are shown.
FIG. 3.
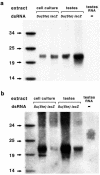
Sizes of short RNAs produced by dsRNA processing in vitro and detected in vivo. (a) Twenty-one- to 23-nt RNA fragments of lacZ and Su(Ste) are produced by dsRNA processing in cell culture and testis extracts. In vitro-synthesized, uniformly labeled lacZ or Su(Ste) dsRNAs were incubated with cell culture or testis extracts, and RNAs were isolated and separated on a 15% denaturing acrylamide gel. Total testis RNA from wild-type males was separated in parallel. 32P-labeled RNA oligonucleotides were used as size markers. (b) The same gel was electroblotted to a membrane and hybridized with a Su(Ste) probe to detect the 25- to 27-nt Su(Ste) RNA in the total testis RNA preparation. Hybridization also increased the strength of signals from the in vitro-synthesized Su(Ste) dsRNA.
FIG. 4.
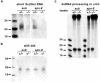
Effects of aub and spn-E mutations on the presence of the short Su(Ste) RNA and of microRNA in vivo and on processing of dsRNA in vitro. (a) Short Su(Ste) RNAs are absent in homozygous aub and spn-E mutants. Equal quantities of total RNA isolated from testes of heterozygous (+/−) and homozygous (−/−) males were separated on a gel and hybridized with a Su(Ste) probe. (b) There is no effect of either mutation on the amount of miR-304. (c) aub and spn-E mutations do not affect in vitro processing of dsRNA to 21- to 23-nt fragments. lacZ dsRNA was incubated with extracts prepared from testes of heterozygous (+/−) or homozygous (−/−) males. No degradation of dsRNA was detected in the absence of testis extract (lysate−).
FIG. 5.
Structures and expression of reporter constructs containing Stellate sequences. (a) The structures of Stellate genes and reporter Ste-lacZ fusion constructs used for Drosophila transformation. The Ste134-lacZ construct contains a 134-bp fragment of the Stellate gene, including 104 bp of nontranscribed sequence followed by 30 bp of the transcribed noncoding region from the first Stellate exon. An arrow indicates the site of substitution of three adjacent nucleotides in the Stellate 5′-UTR sequence in the Ste134mut-lacZ construct. In the β2_tub-Ste-lacZ_ construct, the β2_tub_ promoter drives expression of the Ste open reading frame, detached from intron 2, fused to lacZ. (b) Nucleotide substitutions in the Stellate transcribed region result in relief of Su(Ste)-dependent silencing. β-Gal activity was measured in testis extracts from wild-type (filled bars) and cry1Y (open bars) males for three stocks carrying independent insertions of Stel34-lacZ and eight stocks with independent insertions of Ste134mut-lacZ. There is less difference in lacZ expression between wild-type and cry1Y males that carry the Ste134mut-lacZ construct than between those that carry the Stel34-lacZ construct. Southern analysis confirmed that all of the stocks carry a single transgene insertion. (c) X-Gal staining of testes from transgenic flies carrying the β2_tub-Ste-lacZ_ construct. Weak expression was observed in the germ cells of wild-type males, while in cry1Y males strong staining was detected throughout the testes except in the very tip. cry1Y males have a five- to sevenfold higher level of β-Gal activity than wild-type (XY) males.
Similar articles
- Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline.
Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Aravin AA, et al. Curr Biol. 2001 Jul 10;11(13):1017-27. doi: 10.1016/s0960-9822(01)00299-8. Curr Biol. 2001. PMID: 11470406 - Peculiarities of piRNA-mediated post-transcriptional silencing of Stellate repeats in testes of Drosophila melanogaster.
Kotelnikov RN, Klenov MS, Rozovsky YM, Olenina LV, Kibanov MV, Gvozdev VA. Kotelnikov RN, et al. Nucleic Acids Res. 2009 Jun;37(10):3254-63. doi: 10.1093/nar/gkp167. Epub 2009 Mar 24. Nucleic Acids Res. 2009. PMID: 19321499 Free PMC article. - A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary.
Pélisson A, Sarot E, Payen-Groschêne G, Bucheton A. Pélisson A, et al. J Virol. 2007 Feb;81(4):1951-60. doi: 10.1128/JVI.01980-06. Epub 2006 Nov 29. J Virol. 2007. PMID: 17135323 Free PMC article. - Paralogous stellate and Su(Ste) repeats: evolution and ability to silence a reporter gene.
Gvozdev VA, Kogan GL, Tulin AA, Aravin AA, Naumova NM, Shevelyov YY. Gvozdev VA, et al. Genetica. 2000;109(1-2):131-40. doi: 10.1023/a:1026596419250. Genetica. 2000. PMID: 11293788 Review.
Cited by
- Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways.
Gamez S, Srivastav S, Akbari OS, Lau NC. Gamez S, et al. Cells. 2020 Sep 27;9(10):2180. doi: 10.3390/cells9102180. Cells. 2020. PMID: 32992598 Free PMC article. Review. - Mutations to the piRNA pathway component aubergine enhance meiotic drive of segregation distorter in Drosophila melanogaster.
Gell SL, Reenan RA. Gell SL, et al. Genetics. 2013 Mar;193(3):771-84. doi: 10.1534/genetics.112.147561. Epub 2012 Dec 24. Genetics. 2013. PMID: 23267055 Free PMC article. - Third chromosome candidate genes for conspecific sperm precedence between D. simulans and D. mauritiana.
Levesque L, Brouwers B, Sundararajan V, Civetta A. Levesque L, et al. BMC Genet. 2010 Apr 13;11:21. doi: 10.1186/1471-2156-11-21. BMC Genet. 2010. PMID: 20388218 Free PMC article. - Molecular characterization of embryonic gonads by gene expression profiling in Drosophila melanogaster.
Shigenobu S, Kitadate Y, Noda C, Kobayashi S. Shigenobu S, et al. Proc Natl Acad Sci U S A. 2006 Sep 12;103(37):13728-33. doi: 10.1073/pnas.0603767103. Epub 2006 Sep 1. Proc Natl Acad Sci U S A. 2006. PMID: 16950879 Free PMC article. - Protecting and Diversifying the Germline.
Gleason RJ, Anand A, Kai T, Chen X. Gleason RJ, et al. Genetics. 2018 Feb;208(2):435-471. doi: 10.1534/genetics.117.300208. Genetics. 2018. PMID: 29378808 Free PMC article. Review.
References
- Aravin, A. A., M. Lagos-Quintana, A. Yalcin, M. Zavolan, D. Marks, B. Snyder, T. Gaasterland, J. Meyer, and T. Tuschl. 2003. The Small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337-350. - PubMed
- Aravin, A. A., N. M. Naumova, A. V. Tulin, V. V. Vagin, Y. M. Rozovsky, and V. A. Gvozdev. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11:1017-1027. - PubMed
- Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials