Identification of 315 genes essential for early zebrafish development - PubMed (original) (raw)
Identification of 315 genes essential for early zebrafish development
Adam Amsterdam et al. Proc Natl Acad Sci U S A. 2004.
Abstract
We completed a large insertional mutagenesis screen in zebrafish to identify genes essential for embryonic and early larval development. We isolated 525 mutants, representing lesions in approximately 390 different genes, and we cloned the majority of these. Here we describe 315 mutants and the corresponding genes. Our data suggest that there are roughly 1,400 embryonic-essential genes in the fish. Thus, we have mutations in approximately 25% of these genes and have cloned approximately 22% of them. Re-screens of our collection to identify mutants with specific developmental defects suggest that approximately 50 genes are essential for the development of some individual organs or cell types. Seventy-two percent of the embryonic-essential fish genes have homologues in yeast, 93% have homologues in invertebrates (fly or worm), and 99% have homologues in human. Yeast and worm orthologues of genes that are essential for early zebrafish development have a strong tendency to be essential for viability in yeast and for embryonic development in the worm. Thus, the trait of being a genetically essential gene is conserved in evolution. This mutant collection should be a valuable resource for diverse studies of cell and developmental biology.
Copyright 2004 The National Academy of Sciencs of the USA
Figures
Fig. 1.
Genes essential for zebrafish embryonic development identified by insertional mutagenesis. Genes are listed by mutant number and sorted by evolutionary conservation and gene function. Phenotypic descriptions are available in Table 3, and images are available at
http://web.mit.edu/ccr/pnas\_zebrafish\_mutant\_images/index.html
.
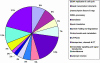
Fig. 2.
Types of genes whose mutation in zebrafish leads to an embryonic visible phenotype. The genes are assigned to the same categories as in Fig. 1 although some categories have been combined.
Fig. 3.
Evolutionary conservation of essential zebrafish genes. Horizonal lines on the Left represent 315 different genes. The genes are listed in the same order as in Fig. 1. In the first four colored columns, the presence of a colored box indicates the presence of one or more homologous genes (
blastp
E value of <10–5) in yeast (either S. cerevisiae or S. pombe), C. elegans, D. melanogaster, or H. sapiens. Green boxes indicate genes without homologues in yeast but with homologues in other SCEs such as Giardia, Plasmodium, Trypanosoma, and/or Chlamydomonas. Thus, yellow boxes represent genes conserved through yeast, green are those found in SCEs other than yeast, red are those found in invertebrates but not SCEs, and blue are vertebrate-specific. The last two genes (with no colored boxes) seem to be fish-specific. The black boxes in the last three columns indicate genes whose mutation leads to one of three phenotypes: cystic kidney, chondrogenesis defects, or reduction or lack of melanocyte pigmentation.
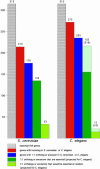
Fig. 4.
Essentialness of genes is evolutionarily conserved. Shown is an analysis of S. cerevisiae (Left) and C. elegans (Right) genes that are homologous to the essential fish genes. In each case, the leftmost columns represent 315 essential fish genes, the red columns show how many of these have homologues in yeast (214) or worm (272), and the blue columns show which have a 1:1 orthologue or “ancestor” gene in yeast (176) or worm (235). The dark green columns represent the number of these yeast or worm orthologues that are essential in their respective species, 135 of the 176 yeast genes, and 155 of the 235 worm genes. The pea green columns show the number that would be predicted to be essential at random, 33 of the 176 yeast genes, and 15 of the 235 worm genes. Thus, the difference between the dark green and pea green columns is the enrichment. In the case of the worm, the pale green extensions to the two green columns represent projections of how many of the orthologues would be found to be essential if 100% of the worm genes had been successfully knocked down by RNAi; this estimate prorates both for the reported failure rate of RNAi and the number of genes for which no RNAi data have been reported. This calculation estimates that 216 of the 235 worm genes are likely to be essential whereas only 22 would be expected to be at random. Note that the percentage of worm orthologues that are essential that is stated in the text does not include the genes for which no RNAi data have been reported.
Similar articles
- Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development.
Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Allende ML, et al. Genes Dev. 1996 Dec 15;10(24):3141-55. doi: 10.1101/gad.10.24.3141. Genes Dev. 1996. PMID: 8985183 - A large-scale insertional mutagenesis screen in zebrafish.
Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. Amsterdam A, et al. Genes Dev. 1999 Oct 15;13(20):2713-24. doi: 10.1101/gad.13.20.2713. Genes Dev. 1999. PMID: 10541557 Free PMC article. - Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development.
Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin SY, Nissen RM, Hopkins N. Golling G, et al. Nat Genet. 2002 Jun;31(2):135-40. doi: 10.1038/ng896. Epub 2002 May 13. Nat Genet. 2002. PMID: 12006978 - Insertional mutagenesis in zebrafish: genes for development, genes for disease.
Amsterdam A. Amsterdam A. Brief Funct Genomic Proteomic. 2006 Mar;5(1):19-23. doi: 10.1093/bfgp/ell008. Epub 2006 Feb 22. Brief Funct Genomic Proteomic. 2006. PMID: 16769673 Review. - "Physiological genomics": mutant screens in zebrafish.
Warren KS, Fishman MC. Warren KS, et al. Am J Physiol. 1998 Jul;275(1):H1-7. doi: 10.1152/ajpheart.1998.275.1.H1. Am J Physiol. 1998. PMID: 9688889 Review.
Cited by
- Ccdc94 protects cells from ionizing radiation by inhibiting the expression of p53.
Sorrells S, Carbonneau S, Harrington E, Chen AT, Hast B, Milash B, Pyati U, Major MB, Zhou Y, Zon LI, Stewart RA, Look AT, Jette C. Sorrells S, et al. PLoS Genet. 2012;8(8):e1002922. doi: 10.1371/journal.pgen.1002922. Epub 2012 Aug 30. PLoS Genet. 2012. PMID: 22952453 Free PMC article. - The Function of V-ATPases in Cancer.
Stransky L, Cotter K, Forgac M. Stransky L, et al. Physiol Rev. 2016 Jul;96(3):1071-91. doi: 10.1152/physrev.00035.2015. Physiol Rev. 2016. PMID: 27335445 Free PMC article. Review. - In vivo chemical screening for modulators of hematopoiesis and hematological diseases.
Zhang Y, Yeh JR. Zhang Y, et al. Adv Hematol. 2012;2012:851674. doi: 10.1155/2012/851674. Epub 2012 Jun 19. Adv Hematol. 2012. PMID: 22778745 Free PMC article. - Evolutionary and developmental analysis reveals KANK genes were co-opted for vertebrate vascular development.
Hensley MR, Cui Z, Chua RF, Simpson S, Shammas NL, Yang JY, Leung YF, Zhang G. Hensley MR, et al. Sci Rep. 2016 Jun 13;6:27816. doi: 10.1038/srep27816. Sci Rep. 2016. PMID: 27292017 Free PMC article. - Mitotic cell rounding and epithelial thinning regulate lumen growth and shape.
Hoijman E, Rubbini D, Colombelli J, Alsina B. Hoijman E, et al. Nat Commun. 2015 Jun 16;6:7355. doi: 10.1038/ncomms8355. Nat Commun. 2015. PMID: 26077034
References
- Golling, G., Amsterdam, A., Sun, Z., Antonelli, M., Maldonado, E., Chen, W., Burgess, S., Haldi, M., Artzt, K, Farrington, S., et al. (2002) Nat. Genet. 31, 135–140. - PubMed
- Amsterdam, A. (2003) Dev. Dyn. 228, 523–534. - PubMed
- Tatusov, R. L., Koonin, E. V. & Lipman D. J. (1997) Science 278, 631–637. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases