Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs - PubMed (original) (raw)
Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs
Ashish Lal et al. EMBO J. 2004.
Abstract
RNA-binding proteins HuR and AUF1 bind to many common AU-rich target mRNAs and exert opposing influence on target mRNA stability, but the functional interactions between HuR and AUF1 have not been systematically studied. Here, using common target RNAs encoding p21 and cyclin D1, we provide evidence that HuR and AUF1 can bind target transcripts on both distinct, nonoverlapping sites, and on common sites in a competitive fashion. In the nucleus, both proteins were found together within stable ribonucleoprotein complexes; in the cytoplasm, HuR and AUF1 were found to bind to target mRNAs individually, HuR colocalizing with the translational apparatus and AUF1 with the exosome. Our results indicate that the composition and fate (stability, translation) of HuR- and/or AUF1-containing ribonucleoprotein complexes depend on the target mRNA of interest, RNA-binding protein abundance, stress condition, and subcellular compartment.
Figures
Figure 1
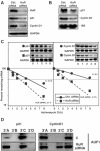
Identification of HuR and AUF1 target mRNAs. (A) RNAs bound to either HuR or AUF1 in HeLa whole-cell lysates were isolated by IP assay using the corresponding antibodies, and the reverse-transcribed radiolabeled products were used for cDNA array hybridization (Materials and methods). Control IP reactions were performed using IgG1. Representative fields of each array are shown (black arrows, specific signals enriched in samples obtained by either HuR IP or AUF1 IP; white arrows, signals enriched in both HuR and AUF1 IP materials). (B) Partial list of genes encoding transcripts that were significantly enriched in the HuR IP only (left), in the AUF1 IP only (center), and in both the HuR and AUF1 IPs (right, left value corresponds to Z ratio for HuR, right value for AUF1). Parentheses, Z ratios. Complete array data are available (
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1361
). (C) For validation of HuR and AUF1 target mRNAs, HeLa whole-cell lysates were prepared and endogenous target transcripts were detected by RT–PCR assay of the IP material. PCR products were visualized on 1% agarose gels. Amplification of housekeeping transcripts encoding GAPDH and SDHA, bound at low levels with the IP material, showed equal loading of IP samples.
Figure 2
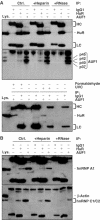
Joint presence of HuR and AUF1 within the same RNP complexes. IP assays were carried out using whole-cell lysate from HeLa cells and either anti-HuR antibody, anti-AUF1 antibody, or IgG1. IP reactions were performed without further treatment (Ctrl.), or in the presence of either heparin (+Heparin) or RNases (+RNase). (A) The presence of HuR (top) and AUF1 (four isoforms, middle) in the IP materials was monitored by Western blotting. Lys., 10 μg of whole-cell lysate, included as Western blotting control; HC, heavy chain; LC, light chain. Bottom: RNPs were crosslinked in intact cells by using either UVC irradiation or formaldehyde (Supplementary material), then subjected to IP and Western blotting. (B) The levels of hnRNP A1 and hnRNP C1/C2 in the IP materials were assessed by Western blotting.
Figure 3
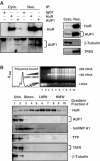
Subcellular distribution of HuR and AUF1. (A) Left: the relative abundance of HuR and AUF1 in RNP complexes obtained through IP (described in the legend of Figure 2) was assessed using either cytoplasmic (50 μg, Cyto.) or nuclear (10 μg, Nuc.) lysates. The relative abundance of HuR, AUF1, β-tubulin (a cytoplasm-specific protein), and TFIID (a nucleus-specific protein) was assessed by Western blotting using 10 μg of either cytoplasmic or nuclear lysates. (B) Representative polysome distribution profile (top left) and pattern of ethidium bromide-stained RNA (top right) from sucrose gradients (Materials and methods). From left to right: fractions lacked ribosomes or ribosome subunits (Unb.), contained ribosome subunits or single ribosomes (monosomes, Mono.), and spanned low-molecular-weight (LMW) and high-molecular-weight (HMW) polysomes. The levels of HuR, AUF1, hnRNP A1, TTP, TIAR, and β-tubulin were monitored by Western blotting using equal volumes of lysates from each fraction.
Figure 4
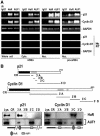
Binding of HuR and AUF1 to distinct sites of the p21 and cyclin D1 mRNAs. (A) Following IP reactions using either whole-cell (1.5 mg, left), cytoplasmic (400 μg, Cyto., center), or nuclear (500 μg, Nuc., right) lysates and anti-HuR, anti-AUF1, or IgG1 antibodies, the binding of endogenous HuR and AUF1 to endogenous target mRNAs (mRNA) was detected by RT–PCR as explained above (Figure 1C); pre-mRNAs (pre-mRNA) were detected by IP of nuclear lysate followed by RT using random hexamers and PCR amplification using primers specific to intron sequences of the p21 and cyclin D1 genes; no amplification was seen in ‘No RT' control samples. (B) Pull-down assays to assess the ability of endogenous HuR and AUF1 to bind to biotinylated transcripts spanning the p21 and cyclin D1 mRNAs. The indicated biotinylated transcripts (1 μg each) were incubated with 40 μg of HeLa whole-cell lysate, whereupon their association with HuR or AUF1 was detected by Western blotting. Lys., 5 μg of whole-cell lysate; CR, coding region. Blackened box, CR; shaded, AREs. Bottom: schematic of proposed binding regions for HuR and AUF1.
Figure 5
Effect of UVC irradiation on the expression of p21 and cyclin D1 mRNAs. At 5 h after exposure of HeLa cells to 15 J/m2 UVC, p21 and cyclin D1 mRNA abundance was assessed by Northern blotting (20 μg total RNA per lane) (A), the presence of HuR and AUF1 on common RNP complexes was monitored by IP and Western blot assays using cytoplasmic lysate, as described above (Figure 2) (B), and the presence of endogenous p21 and cyclin D1 mRNAs with each protein was assayed in the IP material from cytoplasmic lysates by real-time RT–PCR (C); fold differences in abundance were calculated after estimating the _C_T values (representing the number of PCR cycles required to reach a threshold set arbitrarily at 0.8) for each amplification curve (Materials and methods). Low-level amplification of GAPDH and SDHA served to monitor equal addition of RNA from IP materials. (D) Cytoplasmic lysates from either untreated (Ctrl.) or UVC-treated HeLa cultures were fractionated through sucrose gradients, whereupon aliquots from each fraction were used for Western blotting to detect HuR, AUF1, the exosome protein hRrp4p, and the control protein β-tubulin. (E) The relative presence of p21 and cyclin D1 mRNAs in each fraction from the sucrose gradient was determined by Northern blot analysis. (F) The relative abundance of p21 and cyclin D1 mRNAs bound to either HuR or AUF1 in pooled unbound fractions (1 and 2, Unb.) or pooled polysomal fractions (6–10, Polysome) from either untreated or UVC-treated cells was determined by IP+RT–(real-time)PCR.
Figure 6
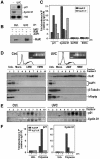
Effect of siRNA-mediated suppression of HuR levels on the expression and stability of p21 and cyclin D1 mRNAs. At 3 days after transfection with an siRNA that suppressed HuR protein levels by RNAi (HuR siRNA) or a control siRNA (C), the expression levels of HuR, cyclin D1, p21, and GAPDH (a control protein serving to monitor the equal loading and transfer of samples) were examined by Western blotting (A), and the abundance of p21 and cyclin D1 mRNAs (and control 18S rRNA) was assessed by Northern blotting (B). (C) A three days after transfection, the half-lives of p21 and cyclin D1 mRNAs in each siRNA group were assessed by using actinomycin D (2 μg/ml); mRNA half-lives (parentheses) were calculated from Northern blotting data (Materials and methods). Insets, representative Northerns, including signals of a stable mRNA encoding GAPDH; 18S signals revealed even loading of samples (not shown). Two independent Northern blotting experiments, yielding comparable results, were performed in order to calculate all mRNA half-lives. (D) The indicated biotinylated transcripts were incubated with 40 μg of lysates prepared from either control-transfected cells (C) or HuR siRNA-transfected cells expressing reduced HuR levels. Pull-down assays to assess the ability of endogenous AUF1 to bind biotinylated transcripts spanning the p21 and cyclin D1 mRNAs were performed as described in Figure 4B.
Figure 7
Effect of siRNA-mediated suppression of AUF1 levels on the expression and stability of p21 and cyclin D1 mRNAs. At 3 days after transfection with plasmid pSILENCER-AUF15, expressing a transcript capable of suppressing AUF1 abundance by RNAi (AUF1 siRNA), or the control plasmid (V), the levels of AUF1, cyclin D1, p21, and GAPDH were examined by Western blotting (A), and the abundance of p21 and cyclin D1 mRNAs (and control 18S rRNA) was assessed by Northern blotting (B). (C) At 3 days after transfection, the half-lives of p21 and cyclin D1 mRNAs in each siRNA group were assessed by using actinomycin D (2 μg/ml); mRNA half-lives (parentheses) were calculated from Northern blotting data that were processed as described in the legend of Figure 6. (D) The indicated biotinylated transcripts were incubated with 40 μg of lysates prepared from either control-transfected HeLa cells (C) or AUF1 siRNA-transfected cells expressing reduced AUF1 levels. Pull-down assays to assess the ability of endogenous HuR to bind biotinylated transcripts spanning the p21 and cyclin D1 mRNAs were performed as described in Figure 4B.
Figure 8
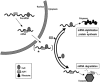
Schematic of the proposed model of HuR and AUF1 physical and functional interaction. See text for details.
Similar articles
- Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3' untranslated region to HuR and AUF1.
Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY. Zou T, et al. Mol Cell Biol. 2010 Nov;30(21):5021-32. doi: 10.1128/MCB.00807-10. Epub 2010 Aug 30. Mol Cell Biol. 2010. PMID: 20805360 Free PMC article. - HuR/methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis.
Vázquez-Chantada M, Fernández-Ramos D, Embade N, Martínez-Lopez N, Varela-Rey M, Woodhoo A, Luka Z, Wagner C, Anglim PP, Finnell RH, Caballería J, Laird-Offringa IA, Gorospe M, Lu SC, Mato JM, Martínez-Chantar ML. Vázquez-Chantada M, et al. Gastroenterology. 2010 May;138(5):1943-53. doi: 10.1053/j.gastro.2010.01.032. Epub 2010 Jan 25. Gastroenterology. 2010. PMID: 20102719 Free PMC article. - FRET-detectable interactions between the ARE binding proteins, HuR and p37AUF1.
David PS, Tanveer R, Port JD. David PS, et al. RNA. 2007 Sep;13(9):1453-68. doi: 10.1261/rna.501707. Epub 2007 Jul 12. RNA. 2007. PMID: 17626845 Free PMC article. - Physiological networks and disease functions of RNA-binding protein AUF1.
Moore AE, Chenette DM, Larkin LC, Schneider RJ. Moore AE, et al. Wiley Interdiscip Rev RNA. 2014 Jul-Aug;5(4):549-64. doi: 10.1002/wrna.1230. Epub 2014 Mar 28. Wiley Interdiscip Rev RNA. 2014. PMID: 24687816 Review. - The eukaryotic translation initiation factor 4E (eIF4E) and HuR RNA operons collaboratively regulate the expression of survival and proliferative genes.
Topisirovic I, Siddiqui N, Borden KL. Topisirovic I, et al. Cell Cycle. 2009 Apr 1;8(7):960-1. Epub 2009 Apr 9. Cell Cycle. 2009. PMID: 19287207 Review. No abstract available.
Cited by
- HuR mediates the synergistic effects of angiotensin II and IL-1β on vascular COX-2 expression and cell migration.
Aguado A, Rodríguez C, Martínez-Revelles S, Avendaño MS, Zhenyukh O, Orriols M, Martínez-González J, Alonso MJ, Briones AM, Dixon DA, Salaices M. Aguado A, et al. Br J Pharmacol. 2015 Jun;172(12):3028-42. doi: 10.1111/bph.13103. Epub 2015 Mar 27. Br J Pharmacol. 2015. PMID: 25653183 Free PMC article. - The tRNA methyltransferase NSun2 stabilizes p16INK⁴ mRNA by methylating the 3'-untranslated region of p16.
Zhang X, Liu Z, Yi J, Tang H, Xing J, Yu M, Tong T, Shang Y, Gorospe M, Wang W. Zhang X, et al. Nat Commun. 2012 Mar 6;3:712. doi: 10.1038/ncomms1692. Nat Commun. 2012. PMID: 22395603 Free PMC article. - The impact of post-transcriptional regulation in the p53 network.
Freeman JA, Espinosa JM. Freeman JA, et al. Brief Funct Genomics. 2013 Jan;12(1):46-57. doi: 10.1093/bfgp/els058. Epub 2012 Dec 14. Brief Funct Genomics. 2013. PMID: 23242178 Free PMC article. Review. - Post-transcriptional regulation of DNA damage-responsive gene expression.
McKay BC. McKay BC. Antioxid Redox Signal. 2014 Feb 1;20(4):640-54. doi: 10.1089/ars.2013.5523. Epub 2013 Sep 12. Antioxid Redox Signal. 2014. PMID: 23905704 Free PMC article. Review. - Beta-Catenin stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich elements of 3'-UTR.
Lee HK, Jeong S. Lee HK, et al. Nucleic Acids Res. 2006;34(19):5705-14. doi: 10.1093/nar/gkl698. Epub 2006 Oct 12. Nucleic Acids Res. 2006. PMID: 17040897 Free PMC article.
References
- Atasoy U, Watson J, Patel D, Keene JD (1998) ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci 111: 3145–3156 - PubMed
- Briata P, Ilengo C, Corte G, Moroni C, Rosenfeld MG, Chen CY, Gherzi R (2003) The Wnt/beta-catenin → Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol Cell 12: 1201–1211 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous