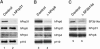RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies - PubMed (original) (raw)
RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies
Nina Schaffert et al. EMBO J. 2004.
Abstract
Cajal bodies (CBs) are subnuclear organelles of animal and plant cells. A role of CBs in the assembly and maturation of small nuclear ribonucleoproteins (snRNP) has been proposed but is poorly understood. Here we have addressed the question where U4/U6.U5 tri-snRNP assembly occurs in the nucleus. The U4/U6.U5 tri-snRNP is a central unit of the spliceosome and must be re-formed from its components after each round of splicing. By combining RNAi and biochemical methods, we demonstrate that, after knockdown of the U4/U6-specific hPrp31 (61 K) or the U5-specific hPrp6 (102 K) protein in HeLa cells, tri-snRNP formation is inhibited and stable U5 mono-snRNPs and U4/U6 di-snRNPs containing U4/U6 proteins and the U4/U6 recycling factor p110 accumulate. Thus, hPrp31 and hPrp6 form an essential connection between the U4/U6 and U5 snRNPs in vivo. Using fluorescence microscopy, we show that, in the absence of either hPrp31 or hPrp6, U4/U6 di-snRNPs as well as p110 accumulate in Cajal bodies. In contrast, U5 snRNPs largely remain in nucleoplasmic speckles. Our data support the idea that CBs may play a role in tri-snRNP recycling.
Figures
Figure 1
Suppression (knockdown) of spliceosomal proteins by siRNAs. (A) A Western blot showing levels of hPrp31, hPrp6, hPrp4 and p110 protein in the nuclear extract prepared from HeLa cells 48 h after transfection with hPrp31-specific siRNA (EA1) and a firefly luciferase siRNA control. (B) A similar blot made 48 h after transfection with hPrp6-specific siRNA (AL5). (C) A Western blot showing protein levels of hPrp31, hPrp6 and SF3b14a protein 48 h after transfection with the SF3b14a-specific siRNA (AS1). The same amounts of protein were loaded from nuclear extracts generated from control and knockdown cells.
Figure 2
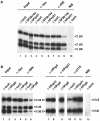
Disruption of the tri-snRNP in vivo by selective depletion of either the U4/U6 protein hPrp31 or the U5 protein hPrp6. (A) Anti-Sm and anti-40K antibodies (denoted α-Sm, α-40 K, etc.) were used to immunoprecipitate snRNPs from the nuclear extract prepared from HeLa cells 48 h after transfection with siRNAs targeting either luciferase, the U2 protein SF3b14a (AS1, ΔSF3b14a) or the U4/U6 protein hPrp31 (EA1, ΔhPrp31). NIS: nonimmune serum from α-40K. Separate controls with non-transfected cells (not shown) revealed patterns identical to those with luciferase-transfected cells. (B) Anti-Sm, anti-40K, anti-hPrp4, anti-hPrp31 and anti-p110 antibodies were used to immunoprecipitate snRNPs from the nuclear extract prepared from HeLa cells 48 h after transfection with siRNAs targeting luciferase, the U4/U6 protein hPrp31 (ΔhPrp31) or the U5 protein hPrp6 (AL5, ΔhPrp6). NIS: non-immune serum from α-hPrp4. In all experiments, RNA was isolated from the immunoprecipitated snRNPs and characterised by Northern blot analysis.
Figure 3
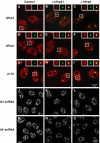
Inhibition of tri-snRNP formation leads to an accumulation of U4/U6-specific components in the Cajal bodies. HeLa cells transfected with siRNA targeting luciferase (control), the U4/U6 protein hPrp31 (EA1, ΔhPrp31) or the U5 protein hPrp6 (AL5, ΔhPrp6) were examined after 48 h using indirect immunofluorescence (A–I) or fluorescence in situ hybridisation (J–O). (A–I) Green, monoclonal antibodies against coilin (showing the Cajal bodies); red, antibodies against the protein in question. (A–C) Transfected HeLa cells were stained with antibodies against coilin (green) and against the U4/U6 protein hPrp4 (red). The accumulation of protein hPrp4 in the Cajal bodies is indicated by the superposition of these (yellow). (D–F) As (A–C), using antibodies against the protein hPrp3 instead of protein hPrp4. (G–I) As above, but using affinity-purified antibodies against protein p110. The red and green fluorescence signals were recorded independently and combined to give the overlay images shown; the insets show separate images for selected Cajal bodies. (J–O) For visualisation of snRNAs, cells were hybridised with Cy3-labelled oligonucleotides complementary to (J–L) U4 snRNA or (M–O) U6 snRNA. For optimal display, contrast and brightness of the images were adjusted.
Figure 4
Sensitivity of the U4/U6 hPrp4 protein distribution pattern to treatment with RNase A. HeLa cells transfected with siRNA targeting the firefly luciferase protein (control), a U4/U6 hPrp31-specific (EA1, ΔhPrp31) or a U5 hPrp6-specific siRNA (AL5, ΔhPrp6) were treated either with (D–F, J–L, P–R) or without RNase A (A–C, G–I, M–O) 48 h after transfection before immunofluorescent investigation of hPrp4 (A–F), p110 distribution in the nucleus (G–L) or U6 snRNA as a control for RNase A digestion (M–R).
Figure 5
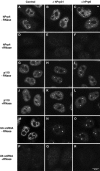
Inhibition of tri-snRNP formation does not lead to an accumulation of U5-specific components in Cajal bodies. HeLa cells transfected with an unspecific siRNA targeting the firefly luciferase protein (control), a U4/U6 hPrp31-specific (EA1, hPrp31) or a U5 hPrp6-specific siRNA (AL5, hPrp6) were analysed after 48 h using immunofluorescence (A–C) or fluorescence in situ hybridisation (D–L). (A–C) Transfected HeLa cells that were stained with antibodies against coilin (green) and against the U5 protein hSnu114 (red). The red and green fluorescence signals were recorded independently and combined to give the overlay images shown; the insets show separate images for selected Cajal bodies. For visualisation of snRNAs, cells were hybridised with fluorescent-labelled oligonucleotides complementary to (D–F) U5 snRNA, (G–I) U2 snRNA or (J–L) U1 snRNA. In rows 2 and 3, the same cells are shown, hybridised with both a Cy3-labelled U5 probe and an Alexa 488-labelled U2 probe to identify the Cajal bodies for quantitative measurements.
Similar articles
- The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation.
Laggerbauer B, Liu S, Makarov E, Vornlocher HP, Makarova O, Ingelfinger D, Achsel T, Lührmann R. Laggerbauer B, et al. RNA. 2005 May;11(5):598-608. doi: 10.1261/rna.2300805. RNA. 2005. PMID: 15840814 Free PMC article. - The assembly of a spliceosomal small nuclear ribonucleoprotein particle.
Patel SB, Bellini M. Patel SB, et al. Nucleic Acids Res. 2008 Nov;36(20):6482-93. doi: 10.1093/nar/gkn658. Epub 2008 Oct 14. Nucleic Acids Res. 2008. PMID: 18854356 Free PMC article. Review. - Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing.
Makarova OV, Makarov EM, Liu S, Vornlocher HP, Lührmann R. Makarova OV, et al. EMBO J. 2002 Mar 1;21(5):1148-57. doi: 10.1093/emboj/21.5.1148. EMBO J. 2002. PMID: 11867543 Free PMC article. - The 20kD protein of human [U4/U6.U5] tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60kD and 90kD proteins.
Teigelkamp S, Achsel T, Mundt C, Göthel SF, Cronshagen U, Lane WS, Marahiel M, Lührmann R. Teigelkamp S, et al. RNA. 1998 Feb;4(2):127-41. RNA. 1998. PMID: 9570313 Free PMC article. - Roles of the U5 snRNP in spliceosome dynamics and catalysis.
Turner IA, Norman CM, Churcher MJ, Newman AJ. Turner IA, et al. Biochem Soc Trans. 2004 Dec;32(Pt 6):928-31. doi: 10.1042/BST0320928. Biochem Soc Trans. 2004. PMID: 15506927 Review.
Cited by
- The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation.
Laggerbauer B, Liu S, Makarov E, Vornlocher HP, Makarova O, Ingelfinger D, Achsel T, Lührmann R. Laggerbauer B, et al. RNA. 2005 May;11(5):598-608. doi: 10.1261/rna.2300805. RNA. 2005. PMID: 15840814 Free PMC article. - In Silico Identification of Hub Genes as Observing Biomarkers for Gastric Cancer Metastasis.
Ajucarmelprecilla A, Pandi J, Dhandapani R, Ramanathan S, Chinnappan J, Paramasivam R, Thangavelu S, Mohammed Ghilan AK, Aljohani SAS, Oyouni AAA, Farasani A, Altayar MA, Althagafi HAE, Alzahrani OR, Durairaj K, Shrestha A. Ajucarmelprecilla A, et al. Evid Based Complement Alternat Med. 2022 Apr 30;2022:6316158. doi: 10.1155/2022/6316158. eCollection 2022. Evid Based Complement Alternat Med. 2022. PMID: 35535159 Free PMC article. Retracted. - The assembly of a spliceosomal small nuclear ribonucleoprotein particle.
Patel SB, Bellini M. Patel SB, et al. Nucleic Acids Res. 2008 Nov;36(20):6482-93. doi: 10.1093/nar/gkn658. Epub 2008 Oct 14. Nucleic Acids Res. 2008. PMID: 18854356 Free PMC article. Review. - Tinker, Tailor, Tumour Suppressor: The Many Functions of PRP4K.
Habib EB, Mathavarajah S, Dellaire G. Habib EB, et al. Front Genet. 2022 Feb 24;13:839963. doi: 10.3389/fgene.2022.839963. eCollection 2022. Front Genet. 2022. PMID: 35281802 Free PMC article. Review. - Three gene-targeted mouse models of RNA splicing factor RP show late-onset RPE and retinal degeneration.
Graziotto JJ, Farkas MH, Bujakowska K, Deramaudt BM, Zhang Q, Nandrot EF, Inglehearn CF, Bhattacharya SS, Pierce EA. Graziotto JJ, et al. Invest Ophthalmol Vis Sci. 2011 Jan 5;52(1):190-8. doi: 10.1167/iovs.10-5194. Print 2011 Jan. Invest Ophthalmol Vis Sci. 2011. PMID: 20811066 Free PMC article.
References
- Achsel T, Ahrens K, Brahms H, Teigelkamp S, Lührmann R (1998) The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol Cell Biol 18: 6756–6766 - PMC - PubMed
- Brow DA (2002) Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 - PubMed
- Burge CB, Tuschl T, Sharp PA (1999) Splicing of precursors to mRNAs by the spliceosomes. In The RNA World, Gesteland RF, Cech TR, Atkins JF (eds), pp 525–560. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases