The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis - PubMed (original) (raw)
The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis
Paul E Staswick et al. Plant Cell. 2004 Aug.
Abstract
Despite its importance in a variety of plant defense responses, our understanding of how jasmonic acid (JA) functions at the biochemical level is limited. Several amino acid conjugates of JA were tested for their ability to complement the JA-insensitive Arabidopsis thaliana mutant jar1-1. Unlike free JA, JA-Ile inhibited root growth in jar1-1 to the same extent as in the wild type, whereas JA-Val, JA-Leu, and JA-Phe were ineffective inhibitors in both genotypes. Thin-layer chromatography and gas chromatography-mass spectrometry (GC-MS) analysis of products produced in vitro by recombinant JAR1 demonstrated that this enzyme forms JA-amido conjugates with several amino acids, including JA-Ile. JA-Val, -Leu, -Ile, and -Phe were each quantified in Arabidopsis seedlings by GC-MS. JA-Ile was found at 29.6 pmole g(-1) fresh weight (FW) in the wild type but was more than sevenfold lower in two jar1 alleles. JA-Leu, -Val, and -Phe were present at only low levels in both genotypes. Expression of wild-type JAR1 in transgenic jar1-1 plants restored sensitivity to JA and elevated JA-Ile to the same level as in the wild type. The ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) conjugated to JA was also found in plant tissue at 18.4 pmole g(-1) FW. JA-ACC was determined not be an effective jasmonate root inhibitor, and surprisingly, was twofold higher in the mutants than in the wild type. This suggests that another JA-conjugating enzyme(s) is present in Arabidopsis. Synthesis of JA-ACC might provide a mechanism to coregulate the availability of JA and ACC for conversion to the active hormones JA-Ile and ethylene, respectively. We conclude that JAR1 is a JA-amino synthetase that is required to activate JA for optimal signaling in Arabidopsis. Plant hormone activation by conjugation to amino acids and the enzymes involved in their formation were previously unknown.
Figures
Figure 1.
Conserved Motifs and Their Relative Position in JAR1 and Bacterial IAA-Lys Synthetase. The remainder of the sequence has no significant similarity. a.a., amino acids.
Figure 2.
Synthesis of Amino Acid Conjugates of JA by Recombinant GST-JAR1. Reactions with JA and each amino acid (single letter abbreviations) were analyzed by TLC and stained for JA with vanillin reagent.
Figure 3.
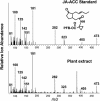
Detection of JA-ACC in Arabidopsis Leaves by GC-MS. The spectra of JA-ACC, the standard eluting at 18.17 min, and the same peak from 6 g of leaf tissue extract are shown. The molecular ion of pentafluorobenzyl (PFB) ester derivative of JA-ACC (473 m/z) and major fragmentation ions are indicated.
Figure 4.
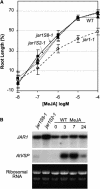
Analysis of jar1-1 Transformants (jar1S2-1 and jar1S8-1) Overexpressing the 35S-JAR1 Construct. (A) Seedling root growth is expressed as a percentage of the same genotype in the absence of JA. Error bars indicate 95% confidence intervals. (B) JAR1 transcript level of transformants and the wild type sprayed with 50 μM MeJA. Each lane was loaded with 9 μg of total leaf RNA from overexpression lines or the wild type. The wild type treated with MeJA was subsequently harvested at times indicated. Hybridization probes were full-length cDNA for JAR1 and AtVSP for the positive control, as indicated. Ribosomal RNA bands stained with ethidium bromide in original gel are shown.
Figure 5.
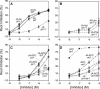
Root Growth Inhibition by JA and Its Amino Acid Conjugates. Inhibition is expressed as percentage untreated for each genotype. Error bars indicate 95% confidence intervals.
Figure 6.
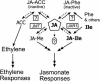
Model for the Role of JAR1 and Related Enzymes in Jasmonate and Ethylene Signaling. The JAR1 (JA-amino synthetase) and JMT are boxed. Other putative JA-amino synthetase(s) are indicated by boxed question marks. Solid and dashed arrows indicate biosynthetic and possible hydrolytic steps, respectively. Double-headed arrows represent signal transduction pathways. The circled question mark indicates uncertainty about the signaling role of free JA. JA-Me, methylester.
Similar articles
- Chemical and genetic exploration of jasmonate biosynthesis and signaling paths.
Kombrink E. Kombrink E. Planta. 2012 Nov;236(5):1351-66. doi: 10.1007/s00425-012-1705-z. Epub 2012 Jul 28. Planta. 2012. PMID: 23011567 Review. - The role of JAR1 in Jasmonoyl-L: -isoleucine production during Arabidopsis wound response.
Suza WP, Staswick PE. Suza WP, et al. Planta. 2008 May;227(6):1221-32. doi: 10.1007/s00425-008-0694-4. Epub 2008 Feb 5. Planta. 2008. PMID: 18247047 - Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone Jasmonoyl-isoleucine for catabolic turnover.
Heitz T, Widemann E, Lugan R, Miesch L, Ullmann P, Désaubry L, Holder E, Grausem B, Kandel S, Miesch M, Werck-Reichhart D, Pinot F. Heitz T, et al. J Biol Chem. 2012 Feb 24;287(9):6296-306. doi: 10.1074/jbc.M111.316364. Epub 2012 Jan 3. J Biol Chem. 2012. PMID: 22215670 Free PMC article. - AtGH3.10 is another jasmonic acid-amido synthetase in Arabidopsis thaliana.
Delfin JC, Kanno Y, Seo M, Kitaoka N, Matsuura H, Tohge T, Shimizu T. Delfin JC, et al. Plant J. 2022 May;110(4):1082-1096. doi: 10.1111/tpj.15724. Epub 2022 Mar 22. Plant J. 2022. PMID: 35247019 - Molecular interaction of jasmonate and phytochrome A signalling.
Hsieh HL, Okamoto H. Hsieh HL, et al. J Exp Bot. 2014 Jun;65(11):2847-57. doi: 10.1093/jxb/eru230. Epub 2014 May 27. J Exp Bot. 2014. PMID: 24868039 Review.
Cited by
- Transcriptomic and volatile signatures associated with maize defense against corn leaf aphid.
Pingault L, Varsani S, Palmer N, Ray S, Williams WP, Luthe DS, Ali JG, Sarath G, Louis J. Pingault L, et al. BMC Plant Biol. 2021 Mar 16;21(1):138. doi: 10.1186/s12870-021-02910-0. BMC Plant Biol. 2021. PMID: 33726668 Free PMC article. - Targeted systems biology profiling of tomato fruit reveals coordination of the Yang cycle and a distinct regulation of ethylene biosynthesis during postclimacteric ripening.
Van de Poel B, Bulens I, Markoula A, Hertog ML, Dreesen R, Wirtz M, Vandoninck S, Oppermann Y, Keulemans J, Hell R, Waelkens E, De Proft MP, Sauter M, Nicolai BM, Geeraerd AH. Van de Poel B, et al. Plant Physiol. 2012 Nov;160(3):1498-514. doi: 10.1104/pp.112.206086. Epub 2012 Sep 13. Plant Physiol. 2012. PMID: 22977280 Free PMC article. - Physiological and Transcriptional Analysis Reveals the Response Mechanism of Camellia vietnamensis Huang to Drought Stress.
Shen S, Yan W, Xie S, Yu J, Yao G, Xia P, Wu Y, Yang H. Shen S, et al. Int J Mol Sci. 2022 Oct 5;23(19):11801. doi: 10.3390/ijms231911801. Int J Mol Sci. 2022. PMID: 36233104 Free PMC article. - Jasmonates: Multifunctional Roles in Stress Tolerance.
Ahmad P, Rasool S, Gul A, Sheikh SA, Akram NA, Ashraf M, Kazi AM, Gucel S. Ahmad P, et al. Front Plant Sci. 2016 Jun 15;7:813. doi: 10.3389/fpls.2016.00813. eCollection 2016. Front Plant Sci. 2016. PMID: 27379115 Free PMC article. Review. - Chemical and genetic exploration of jasmonate biosynthesis and signaling paths.
Kombrink E. Kombrink E. Planta. 2012 Nov;236(5):1351-66. doi: 10.1007/s00425-012-1705-z. Epub 2012 Jul 28. Planta. 2012. PMID: 23011567 Review.
References
- Bleecker, A.B., Estelle, M.A., Somerville, C., and Kende, H. (1988). Insensitivity to ethylene is conferred by a dominant mutation in Arabidopsis thaliana. Science 241, 1086–1089. - PubMed
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. - PubMed
- Conconi, A., Smerdon, M.J., Howe, G.A., and Ryan, C.A. (1996). The octadecanoid signaling pathway in plants mediates a response to ultraviolet radiation. Nature 383, 826–829. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous