Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes - PubMed (original) (raw)
Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes
Robert Kurzbauer et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Signaling pathways in eukaryotic cells are often controlled by the formation of specific signaling complexes, which are coordinated by scaffold and adaptor proteins. Elucidating their molecular architecture is essential to understand the spatial and temporal regulation of cellular signaling. p14 and MP1 form a tight (K(d) = 12.8 nM) endosomal adaptor/scaffold complex, which regulates mitogen-activated protein kinase (MAPK) signaling. Here, we present the 1.9-A crystal structure of a biologically functional p14/MP1 complex. The overall topology of the individual MP1 and p14 proteins is almost identical, having a central five-stranded beta-sheet sandwiched between a two-helix and a one-helix layer. Formation of the p14/MP1 heterodimer proceeds by beta-sheet augmentation and yields a unique, almost symmetrical, complex with several potential protein-binding sites on its surface. Mutational analysis allowed identification of the p14 endosomal adaptor motif, which seems to orient the complex relative to the endosomal membrane. Two highly conserved and hydrophobic protein-binding sites are located on the opposite "cytoplasmic" face of the p14/MP1 heterodimer and might therefore function as docking sites for the target proteins extracellular regulated kinase (ERK) 1 and MAPK/ERK kinase 1. Furthermore, detailed sequence analyses revealed that MP1/p14, together with profilins, define a protein superfamily of small subcellular adaptor proteins, named ProflAP. Taken together, the presented work provides insight into the spatial regulation of MAPK signaling, illustrating how p14 and MP1 collaborate as an endosomal adaptor/scaffold complex.
Figures
Fig. 1.
In vitro properties of the recombinant p14/MP1 complex. (A) SDS/PAGE of the purified p14/MP1 complex. (B) Quantitative Biosensor analysis of MP1 association with p14. The CM5 sensor chip was loaded with GST-tagged p14 and purified MP1 was injected at the indicated concentration (10–250 nM). (C) The recombinant p14/MP1 complex enhances ERK1/2 activation (phosphorylation). Increasing amounts of purified p14/MP1 were added to a cell lysate from EGF-stimulated HeLa cells and incubated at 30°C for 30 min. Cell lysates were separated by SDS/PAGE and probed with the indicated antibodies (α-). (D) Recombinant p14/MP1 binds endosomal membranes in a concentration-dependent manner. (Upper) p14/MP1 complex and GST were incubated with buffer, cytosol, or total cell membranes. Only p14/MP1 copelleted with total cell membranes. (Lower) p14/MP1 complex and GST were incubated with increasing amounts of purified endosomes. Only p14/MP1 copelleted with purified endosomes. Input (before 100,000 × g centrifugation) membrane pellets and supernatant were separated by SDS/PAGE and probed with the indicated antibodies.
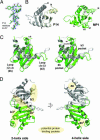
Fig. 2.
Structure of p14/MP1. (A) Experimental electron density map of a representative region of the p14/MP1 complex overlaid with the final model. The map was calculated at 1.9-Å resolution and is contoured at 1.2σ. (B) Ribbon diagram of the individual MP1 (green) and p14 (white) monomers. A color ramp was used to illustrate the course of the polypeptide. All secondary structure elements and the termini are given. (C) Stereoview of the superposition of MP1 (green) and p14 (white). The most divergent regions (Left) and some of the prominent surface pockets (Right) are labeled. (D) Ribbon diagram of the heterodimer with MP1 in green and p14 in white. On the basis of the constituting helix layers, two different faces of the p14/MP1 complex can be distinguished, i.e., the two- and four-helix sides, which are presented in orthogonal mode. The nomenclature and location of possible protein–protein interaction sites are given.
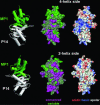
Fig. 3.
Surface properties of the p14/MP1 complex. The four-helix side (Upper) and the two-helix side (Lower) are shown. Left shows Relative orientation of the heterodimer, whereas Center and Right show the p14/MP1 molecular surfaces of the corresponding orientations. (Center) Conservation pattern calculated with
al
2
co
(29) is mapped on the surface, with magenta indicating the most-conserved areas and green indicating the least-conserved areas. (Right) Electrostatic potential calculated with
mead
is mapped on the p14/MP1 surface. Red indicates negatively charged and blue indicates positively charged regions.
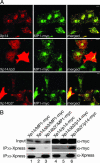
Fig. 4.
Mutations affecting the b3 loop of p14 alter the subcellular distribution of p14/MP1. (A) Xpress-tagged WT p14, p14Δb3, and p14b3* proteins were coexpressed with MP1-myc. Subsequently, cells were subjected to indirect immunofluorescence analysis using anti-Xpress and anti-myc antibodies. Xp14 (red) and MP1-myc (green) colocalize on late endosomes (yellow). Xp14Δb3 was localized in the cytoplasm and did not colocalize with MP1 on late endosomes. Xp14b3* was found to colocalize with MP1-myc on late endosomes but also partially in the cytoplasm. (B) Xp14, Xp14Δb3, and Xp14b3* were coexpressed with MP1-myc or myc-tagged p14. The Xpress-tagged proteins were immunoprecipitated (IP) with an anti-Xpress antibody. To demonstrate equal MP1-myc or myc-p14 expression, cell lysates were probed with anti-myc antibodies (Input). Probing p14 immunoprecipitates with anti-myc (MP1 and p14) antibodies detected interaction of Xp14 with MP1-myc (lane 1) and of Xp14b3* with myc-p14 (lane 6).
Similar articles
- A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment.
Wunderlich W, Fialka I, Teis D, Alpi A, Pfeifer A, Parton RG, Lottspeich F, Huber LA. Wunderlich W, et al. J Cell Biol. 2001 Feb 19;152(4):765-76. doi: 10.1083/jcb.152.4.765. J Cell Biol. 2001. PMID: 11266467 Free PMC article. - The structure of the MAPK scaffold, MP1, bound to its partner, p14. A complex with a critical role in endosomal map kinase signaling.
Lunin VV, Munger C, Wagner J, Ye Z, Cygler M, Sacher M. Lunin VV, et al. J Biol Chem. 2004 May 28;279(22):23422-30. doi: 10.1074/jbc.M401648200. Epub 2004 Mar 11. J Biol Chem. 2004. PMID: 15016825 - Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction.
Teis D, Wunderlich W, Huber LA. Teis D, et al. Dev Cell. 2002 Dec;3(6):803-14. doi: 10.1016/s1534-5807(02)00364-7. Dev Cell. 2002. PMID: 12479806 - Interactions between kinase scaffold MP1/p14 and its endosomal anchoring protein p18.
Magee J, Cygler M. Magee J, et al. Biochemistry. 2011 May 10;50(18):3696-705. doi: 10.1021/bi101972y. Epub 2011 Apr 15. Biochemistry. 2011. PMID: 21452851 - Regulation of ASK1 signaling by scaffold and adaptor proteins.
Rusnak L, Fu H. Rusnak L, et al. Adv Biol Regul. 2017 Dec;66:23-30. doi: 10.1016/j.jbior.2017.10.003. Epub 2017 Oct 16. Adv Biol Regul. 2017. PMID: 29102394 Review.
Cited by
- Molecular characterization and expression profile of MAP2K1ip1/MP1 gene from tiger shrimp, Penaeus monodon.
Yang L, Liu X, Huang J, Yang Q, Qiu L, Liu W, Jiang S. Yang L, et al. Mol Biol Rep. 2012 May;39(5):5811-8. doi: 10.1007/s11033-011-1391-0. Epub 2011 Dec 31. Mol Biol Rep. 2012. PMID: 22209950 - C7orf59/LAMTOR4 phosphorylation and structural flexibility modulate Ragulator assembly.
Rasheed N, Lima TB, Mercaldi GF, Nascimento AFZ, Silva ALS, Righetto GL, Bar-Peled L, Shen K, Sabatini DM, Gozzo FC, Aparicio R, Smetana JHC. Rasheed N, et al. FEBS Open Bio. 2019 Sep;9(9):1589-1602. doi: 10.1002/2211-5463.12700. Epub 2019 Jul 28. FEBS Open Bio. 2019. PMID: 31314152 Free PMC article. - Discovery of new Longin and Roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II.
Levine TP, Daniels RD, Wong LH, Gatta AT, Gerondopoulos A, Barr FA. Levine TP, et al. Small GTPases. 2013 Apr-Jun;4(2):62-9. doi: 10.4161/sgtp.24262. Epub 2013 Mar 19. Small GTPases. 2013. PMID: 23511850 Free PMC article. Review. - Amino acid signalling upstream of mTOR.
Jewell JL, Russell RC, Guan KL. Jewell JL, et al. Nat Rev Mol Cell Biol. 2013 Mar;14(3):133-9. doi: 10.1038/nrm3522. Epub 2013 Jan 30. Nat Rev Mol Cell Biol. 2013. PMID: 23361334 Free PMC article. Review. - Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity.
Araki Y, Ku WC, Akioka M, May AI, Hayashi Y, Arisaka F, Ishihama Y, Ohsumi Y. Araki Y, et al. J Cell Biol. 2013 Oct 28;203(2):299-313. doi: 10.1083/jcb.201304123. J Cell Biol. 2013. PMID: 24165940 Free PMC article.
References
- Pawson, T. & Scott, J. D. (1997) Science 278, 2075–2080. - PubMed
- Morrison, D. K. & Davis, R. J. (2003) Annu. Rev. Cell Dev. Biol. 19, 91–118. - PubMed
- Pawson, T. (2004) Cell 116, 191–203. - PubMed
- Chang, L. & Karin, M. (2001) Nature 410, 37–40. - PubMed
- Garrington, T. P. & Johnson, G. L. (1999) Curr. Opin. Cell Biol. 11, 211–218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous