The gene encoding human retinoic acid-receptor-related orphan receptor alpha is a target for hypoxia-inducible factor 1 - PubMed (original) (raw)
The gene encoding human retinoic acid-receptor-related orphan receptor alpha is a target for hypoxia-inducible factor 1
Caroline Chauvet et al. Biochem J. 2004.
Abstract
Retinoic acid-receptor-related orphan receptor (ROR) alpha is a nuclear receptor involved in many pathophysiological processes such as cerebellar ataxia, inflammation, atherosclerosis and angiogenesis. In the present study we first demonstrate that hypoxia increases the amount of Rora transcripts in a wide panel of cell lines derived from diverse tissues. In addition, we identified a functional promoter sequence upstream of the first exon of the human Rora gene, spanning -487 and -45 from the translation initiation site of RORalpha1. When cloned in a luciferase reporter vector, this sequence allowed the efficient transcription of the luciferase gene in several cell lines. Interestingly, the activity of the Rora promoter was enhanced by hypoxia in HepG2 human hepatoma cells, and this effect was dependent on an HRE (hypoxia response element) spanning from -229 to -225. Using electrophoretic-mobility-shift assays, we showed that HIF-1 (hypoxia-inducible factor 1), which plays a key role in the transcriptional response to hypoxia, bound to this HRE. Overexpression of HIF-1alpha increased the activity of the Rora promoter through the HRE. Overexpression of a dominant-negative form of HIF-1alpha producing transcriptionally inactive HIF-1alpha/HIF-1beta dimers abolished hypoxic activation of the Rora promoter. This indicated that HIF-1 is involved in the response of RORalpha to hypoxia. Taken together, our data reveal Rora as a new HIF-1 target gene. This illustrates, at the molecular level, the existence of cross-talk between signalling pathways mediated by HIF-1 and those mediated by nuclear receptors.
Figures
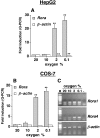
Figure 1. Both the Rora1 and Rora4 mRNA levels are increased under hypoxic conditions
HepG2 cells and COS-7 cells were cultured for 24 h under normoxia (20% O2) or hypoxia (10, 2, or 0.1% O2). (A and B) Total RNA was analysed for Rora mRNA (all isoforms) by quantitative RT-PCR. β-Actin mRNA was monitored as a control. Results are expressed relative to the values at 20% O2 and are given as means±S.E.M. (_n_=3). Significant differences (P<0.01) from 20%-O2 controls are indicated by **. (C) Total RNA from COS-7 cells was analysed for Rora1 mRNA (35 cycles), Rora4 mRNA (30 cycles) by semi-quantitative RT-PCR. Amplification of β-actin mRNA (17 cycles) was performed as a control. The PCR controls (without cDNA) are shown (lanes −). The photographs are representative of those obtained after at least three independent experiments.
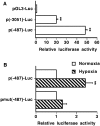
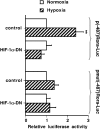
Figure 2. A functional promoter up-regulated by hypoxia lies in upstream region close to the first exon of the human Rora gene
(A) HepG2 cells cultured in 12-well plates were transfected with 500 ng/well of the p(−3051)Rora-Luc reporter vector or with 500 ng/well of the p(−487)Rora-Luc reporter vector, which allow expression of the luciferase gene under the control of the human genomic sequences between nucleotides −3051 and −45, or between nucleotides −487 and −45, from the Rora1 translation initiation site respectively, or with 500 ng/well of the promoter-less pGL3-Luc control vector. The luciferase activity of the p(−3051)Rora-Luc and p(−487)Rora-Luc vectors is expressed relative to that of the promoter-less pGL3-Luc vector. Results are given as means±S.E.M. (_n_=12). Significant differences (P<0.01) from the luciferase activity of the pGL3-Luc vector are indicated by **. (B) HepG2 cells cultured in 60-mm-diameter glass dishes were transfected with 5 μg of the p(−487)Rora-Luc vector or with 5 μg of the pmut(−487)Rora-Luc vector, which bears three point mutations in the putative HRE located between nucleotides −229 and −225 from the Rora1 translation initiation site. The luciferase activities of the reporter vectors in cells cultured for 24 h under hypoxia (2% O2) are expressed relative to their respective activities in control cells cultured under normoxia (20% O2). Results are given as means±S.E.M. (_n_=6). Significant differences (P<0.01) from the luciferase activity of the reporter vectors in cells cultured under normoxia (20% O2) are indicated by **.
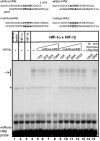
Figure 3. HIF-1α binds specifically to the −229-to-−225 HRE of the Rora promoter
Radiolabelled wt-_Rora_-HRE probe was incubated with 1 μl of HIF-1α programmed lysate (lane 2), 1 μl of HIF-1β programmed lysate (lane 3), or both (lanes 4–15). Unprogrammed reticulocyte lysate was used as a control (lane 1) and to keep the total volume of lysate at 2 μl (lanes 2 and 3). Competition assays were carried out by incubating the radiolabelled wt-_Rora_-HRE probe with the HIF-1α and HIF-1β programmed lysates in the presence of unlabelled wt-_Rora_-HRE (lanes 5–8) or mut_Rora_-HRE (lanes 9–12) double-stranded oligonucleotides at a 10-, 25-, 50- or 100-fold molar excess. A 100-fold molar excess of the wt-_Epo_-HRE (lane 14) or mut-_Epo_-HRE (lane 15) double-stranded oligonucleotides was also used. The arrow points to the specific HIF-1–probe complexes. The asterisk (*) indicates an unrelated complex formed with proteins present in the unprogrammed lysate. Sequences of the oligonucleotides used in the electrophoretic-mobility-shift assays are represented in the upper part of the Figure. The sequence of the HREs is written in bold, the point mutations are
underlined
, and the nucleotides written in lower case do not belong to the Rora or the Epo sequence and were added for convenience.
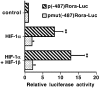
Figure 4. HIF-1 stimulates the activity of the Rora promoter through the −229-to-−225 HRE
HepG2 cells cultured in 12-well plates were transfected with 500 ng/well of the p(−487)Rora-Luc or with 500 ng/well of the pmut(−487)Rora-Luc reporter vector, together with 333 ng/well of the pcDNA3-HIF1α expression vector, and/or with 33 ng/well of the pcDNA3-HIF1β expression vector, or with the pcDNA3 control expression vector. The total amount of transfected DNA was kept constant at 866 ng by adding the pcDNA3 insertless vector. The luciferase activities in the presence of HIF-1 are expressed relative to those in the absence of HIF-1 expression vector. Results are given as means±S.E.M. (_n_=12). Significant differences (P<0.01) from the luciferase activity of the reporter vectors in the absence of HIF-1 expression vector are indicated by **.
Figure 5. A DN form of HIF-1α abolishes the stimulatory effect of hypoxia on the Rora promoter
HepG2 cells cultured in 60-mm-diameter glass dishes were transfected with 5 μg of the p(−487)Rora-Luc or 5 μg of the pmut(−487)Rora-Luc reporter vector, without or with 500 ng of the pcDNA3-HIF1αDN expression vector, while keeping the total amount of transfected DNA constant to 5.5 μg by adding the pcDNA3 insertless vector. The luciferase activities of the reporter vectors in cells cultured for 24 h under hypoxia (2% O2) are expressed relative to their respective activities in control cells cultured under normoxia (20% O2). Results are given as means±S.E.M. (_n_=6). Significant differences (P<0.01) from the luciferase activity of the reporter vectors in normoxia are indicated by **.
Similar articles
- Transcriptional activation of HIF-1 by RORalpha and its role in hypoxia signaling.
Kim EJ, Yoo YG, Yang WK, Lim YS, Na TY, Lee IK, Lee MO. Kim EJ, et al. Arterioscler Thromb Vasc Biol. 2008 Oct;28(10):1796-802. doi: 10.1161/ATVBAHA.108.171546. Epub 2008 Jul 24. Arterioscler Thromb Vasc Biol. 2008. PMID: 18658046 - Hypoxia-induced activation of the retinoic acid receptor-related orphan receptor alpha4 gene by an interaction between hypoxia-inducible factor-1 and Sp1.
Miki N, Ikuta M, Matsui T. Miki N, et al. J Biol Chem. 2004 Apr 9;279(15):15025-31. doi: 10.1074/jbc.M313186200. Epub 2004 Jan 23. J Biol Chem. 2004. PMID: 14742449 - Negative and positive regulation of HIF-1: a complex network.
Bárdos JI, Ashcroft M. Bárdos JI, et al. Biochim Biophys Acta. 2005 Jul 25;1755(2):107-20. doi: 10.1016/j.bbcan.2005.05.001. Biochim Biophys Acta. 2005. PMID: 15994012 Review. - Hypoxia regulation of gene transcription.
Caro J. Caro J. High Alt Med Biol. 2001 Summer;2(2):145-54. doi: 10.1089/152702901750265251. High Alt Med Biol. 2001. PMID: 11442996 Review.
Cited by
- Control of chondrocyte gene expression by actin dynamics: a novel role of cholesterol/Ror-alpha signalling in endochondral bone growth.
Woods A, James CG, Wang G, Dupuis H, Beier F. Woods A, et al. J Cell Mol Med. 2009 Sep;13(9B):3497-516. doi: 10.1111/j.1582-4934.2009.00684.x. J Cell Mol Med. 2009. PMID: 20196782 Free PMC article. - The Relationship Between HIF1α and Clock Gene Expression in Patients with Obstructive Sleep Apnea.
Xie T, Guo D, Luo J, Guo Z, Zhang S, Wang A, Wang X, Wang X, Cao W, Su L, Guo J, Huang R, Xiao Y. Xie T, et al. Nat Sci Sleep. 2022 Mar 8;14:381-392. doi: 10.2147/NSS.S348580. eCollection 2022. Nat Sci Sleep. 2022. PMID: 35299629 Free PMC article. - Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism.
Jetten AM. Jetten AM. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. Epub 2009 Apr 3. Nucl Recept Signal. 2009. PMID: 19381306 Free PMC article. Review. - On the relationship between VDR, RORα and RORγ receptors expression and HIF1-α levels in human melanomas.
Brożyna AA, Jóźwicki W, Jetten AM, Slominski AT. Brożyna AA, et al. Exp Dermatol. 2019 Sep;28(9):1036-1043. doi: 10.1111/exd.14002. Epub 2019 Aug 8. Exp Dermatol. 2019. PMID: 31287590 Free PMC article. - Nuclear receptor RORα regulates pathologic retinal angiogenesis by modulating SOCS3-dependent inflammation.
Sun Y, Liu CH, SanGiovanni JP, Evans LP, Tian KT, Zhang B, Stahl A, Pu WT, Kamenecka TM, Solt LA, Chen J. Sun Y, et al. Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10401-6. doi: 10.1073/pnas.1504387112. Epub 2015 Aug 4. Proc Natl Acad Sci U S A. 2015. PMID: 26243880 Free PMC article.
References
- Semenza G. L. HIF-1 and human disease: one highly involved factor. Genes Dev. 2000;14:1983–1991. - PubMed
- Wenger R. H. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 2002;16:1151–1162. - PubMed
- Pugh C. W., Ratcliffe P. J. Regulation of angiogenesis by hypoxia: role of the HIF system. 2003;9:677–684. - PubMed
- Salceda S., Caro J. Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin–proteasome system under normoxic conditions. J. Biol. Chem. 1997;272:22642–22647. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources