Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes - PubMed (original) (raw)
Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes
Eric Bonnet et al. Proc Natl Acad Sci U S A. 2004.
Erratum in
- Proc Natl Acad Sci U S A. 2005 Mar 22;102(12):4655
Abstract
MicroRNAs (miRNAs) are an extensive class of tiny RNA molecules that regulate the expression of target genes by means of complementary base pair interactions. Although the first miRNAs were discovered in Caenorhabditis elegans, >300 miRNAs were recently documented in animals and plants, both by cloning methods and computational predictions. We present a genome-wide computational approach to detect miRNA genes in the Arabidopsis thaliana genome. Our method is based on the conservation of short sequences between the genomes of Arabidopsis and rice (Oryza sativa) and on properties of the secondary structure of the miRNA precursor. The method was fine-tuned to take into account plant-specific properties, such as the variable length of the miRNA precursor sequences. In total, 91 potential miRNA genes were identified, of which 58 had at least one nearly perfect match with an Arabidopsis mRNA, constituting the potential targets of those miRNAs. In addition to already known transcription factors involved in plant development, the targets also comprised genes involved in several other cellular processes, such as sulfur assimilation and ubiquitin-dependent protein degradation. These findings considerably broaden the scope of miRNA functions in plants.
Figures
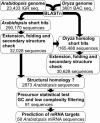
Fig. 1.
Overview of the
mirfinder
computational pipeline to detect miRNAs in plants. For details, see text.
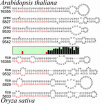
Fig. 2.
RNA secondary structure models of the eight precursor molecules (four each from Arabidopsis and rice) of the miRNAs that target the 5′ UTR of the Arabidopsis gene At2g33770.1 and its rice homolog. The precursor molecules are drawn with the miRNAs aligned and highlighted in red. (Middle) The graph represents the Shannon entropy of the nucleotide content of each corresponding position on the 3′ strand of the precursors. A low value means that all molecules have the same nucleotide at that position, whereas a high value reflects little or no nucleotide conservation. The sequence divergence outside of the mature miRNA positions is clearly shown. RNA sequences are drawn from 5′ to 3′ in clockwise orientation. All precursors are truncated at the same position. RNA secondary structure drawings were made by using
rnaviz
(59).
Similar articles
- Computational identification of novel family members of microRNA genes in Arabidopsis thaliana and Oryza sativa.
Li Y, Li W, Jin YX. Li Y, et al. Acta Biochim Biophys Sin (Shanghai). 2005 Feb;37(2):75-87. Acta Biochim Biophys Sin (Shanghai). 2005. PMID: 15685364 - Computational detection of microRNAs targeting transcription factor genes in Arabidopsis thaliana.
Li X, Zhang YZ. Li X, et al. Comput Biol Chem. 2005 Oct;29(5):360-7. doi: 10.1016/j.compbiolchem.2005.08.005. Epub 2005 Oct 10. Comput Biol Chem. 2005. PMID: 16221572 - Diversity of endogenous small non-coding RNAs in Oryza sativa.
Chen Z, Zhang J, Kong J, Li S, Fu Y, Li S, Zhang H, Li Y, Zhu Y. Chen Z, et al. Genetica. 2006 Sep-Nov;128(1-3):21-31. doi: 10.1007/s10709-005-2486-0. Genetica. 2006. PMID: 17028937 - MicroRNA-mediated signaling involved in plant root development.
Meng Y, Ma X, Chen D, Wu P, Chen M. Meng Y, et al. Biochem Biophys Res Commun. 2010 Mar 12;393(3):345-9. doi: 10.1016/j.bbrc.2010.01.129. Epub 2010 Feb 6. Biochem Biophys Res Commun. 2010. PMID: 20138828 Review. - Computational analysis of miRNA targets in plants: current status and challenges.
Dai X, Zhuang Z, Zhao PX. Dai X, et al. Brief Bioinform. 2011 Mar;12(2):115-21. doi: 10.1093/bib/bbq065. Epub 2010 Sep 21. Brief Bioinform. 2011. PMID: 20858738 Review.
Cited by
- Identification of small RNAs during cold acclimation in Arabidopsis thaliana.
Tiwari B, Habermann K, Arif MA, Weil HL, Garcia-Molina A, Kleine T, Mühlhaus T, Frank W. Tiwari B, et al. BMC Plant Biol. 2020 Jun 29;20(1):298. doi: 10.1186/s12870-020-02511-3. BMC Plant Biol. 2020. PMID: 32600430 Free PMC article. - Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice.
Xie K, Wu C, Xiong L. Xie K, et al. Plant Physiol. 2006 Sep;142(1):280-93. doi: 10.1104/pp.106.084475. Epub 2006 Jul 21. Plant Physiol. 2006. PMID: 16861571 Free PMC article. - Munich information center for protein sequences plant genome resources: a framework for integrative and comparative analyses 1(W).
Schoof H, Spannagl M, Yang L, Ernst R, Gundlach H, Haase D, Haberer G, Mayer KF. Schoof H, et al. Plant Physiol. 2005 Jul;138(3):1301-9. doi: 10.1104/pp.104.059188. Plant Physiol. 2005. PMID: 16010004 Free PMC article. - Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development.
Padmanabhan MS, Goregaoker SP, Golem S, Shiferaw H, Culver JN. Padmanabhan MS, et al. J Virol. 2005 Feb;79(4):2549-58. doi: 10.1128/JVI.79.4.2549-2558.2005. J Virol. 2005. PMID: 15681455 Free PMC article. - Identification of miRNA-mRNA Regulatory Modules Involved in Lipid Metabolism and Seed Development in a Woody Oil Tree (Camellia oleifera).
Wu B, Ruan C, Shah AH, Li D, Li H, Ding J, Li J, Du W. Wu B, et al. Cells. 2021 Dec 27;11(1):71. doi: 10.3390/cells11010071. Cells. 2021. PMID: 35011633 Free PMC article.
References
- Bartel, D. P. (2004) Cell 116, 281-297. - PubMed
- Carrington, J. C. & Ambros, V. (2003) Science 301, 336-338. - PubMed
- Lai, E. C. (2003) Curr. Biol. 13, R925-R936. - PubMed
- Lee, R. C., Feinbaum, R. L. & Ambros, V. (1993) Cell 75, 843-854. - PubMed
- Wightman, B., Ha, I. & Ruvkun, G. (1993) Cell 75, 855-862. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases