Model for the interaction of gammaherpesvirus 68 RING-CH finger protein mK3 with major histocompatibility complex class I and the peptide-loading complex - PubMed (original) (raw)
Model for the interaction of gammaherpesvirus 68 RING-CH finger protein mK3 with major histocompatibility complex class I and the peptide-loading complex
Xiaoli Wang et al. J Virol. 2004 Aug.
Abstract
The mK3 protein of gammaherpesvirus 68 and the kK5 protein of Kaposi's sarcoma-associated herpesvirus are members of a family of structurally related viral immune evasion molecules that all possess a RING-CH domain with ubiquitin ligase activity. These proteins modulate the expression of major histocompatibility complex class I molecules (mK3 and kK5) as well as other molecules like ICAM-1 and B7.2 (kK5). Previously, mK3 was shown to ubiquitinate nascent class I molecules, resulting in their rapid degradation, and this process was found to be dependent on TAP and tapasin, endoplasmic reticulum molecules involved in class I assembly. Here, we demonstrate that in murine cells, kK5 does not affect class I expression but does downregulate human B7.2 molecules in a TAP/tapasin-independent manner. These differences in substrate specificity and TAP/tapasin dependence between mK3 and kK5 permitted us, using chimeric molecules, to map the sites of mK3 interaction with TAP/tapasin and to determine the requirements for substrate recognition by mK3. Our findings indicate that mK3 interacts with TAP1 and -2 via their C-terminal domains and with class I molecules via their N-terminal domains. Furthermore, by orienting the RING-CH domain of mK3 appropriately with respect to class I, mK3 binding to TAP/tapasin, rather than the presence of unique sequences in class I, appears to be the primary determinant of substrate specificity.
Figures
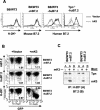
FIG. 1.
kK5 but not mK3 downregulates human B7.2 in mouse cells in a tapasin-independent manner. (A) wt (B6/WT3) and tapasin-deficient fibroblasts (Tpn−/−) (both H-2b) were first transduced with either human or mouse B7.2, followed by transduction with pMSCV-IRES-GFP (pMIG; vector) only or pMIG-kK5. Stable lines were analyzed for surface expression of human or mouse B7.2, or Kb, as indicated. The thick solid lines indicate staining of vector-only cells; dashed lines indicate staining of kK5-expressing cells. (B) wt cells expressing either mouse B7.2 (mB7.2) or human B7.2 (huB7.2) were transduced with pMIG (vector) only or pMIG-mK3 (mK3). After culture for 24 h with 100 U of IFN-γ/ml, surface B7.2 or Kb expression (y axis) versus GFP fluorescence (x axis) was monitored. The GFP+ and GFP− populations in each transductant represent the cells transduced or not transduced by pMIG-containing virus, respectively. The number in each plot represents the ratio of B7.2 or Kb staining between GFP+ and GFP− populations. (C) Digitonin cell lysates from the cells used in panel B were immunoprecipitated with MAb 28-14-8 for H-2Db (A) or GL1/FUN-1 for mouse/human B7.2 (B). Precipitates were then blotted with the antibody listed.
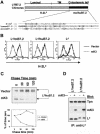
FIG. 2.
Ld/B7.2 chimeras are sensitive to mK3, and this correlates with their ability to associate with TAP/tapasin. (A) Schematic representation of Ld/B7.2 chimeras constructed with the luminal and TM regions of H-2Ld and cytoplasmic tail of either mouse B7.2 (Ld/mB7.2) or human B7.2 (Ld/huB7.2). A sequence alignment of the cytoplasmic tails of the indicated proteins is shown below the diagram. Lysine residues (K) are shown in bold. Identical or similar residues are shaded. (B) Constructs in panel A were introduced into B6/WT3 cells, followed by transduction with pMIG only (vector) or pMIG-mK3 (mK3). Sorting was used to enrich for GFP+ cells. Cells were incubated for 24 h with 100 U of IFN-γ/ml and then analyzed for expression of wt Ld or the Ld/B7.2 constructs by using MAb 30-5-7. Solid lines, staining of vector-only cells; dashed lines, staining of the mK3-expressing cells. (C) IFN-γ-treated cells were pulse-labeled with [35S]Cys/Met and chased for the indicated times with unlabeled Cys/Met. Ld/huB7.2 molecules were precipitated with MAb 64-3-7. Precipitates were resolved by SDS-PAGE and visualized by autoradiography. Ld/huB7.2 bands are indicated (upper panel). In the lower panel, relative band intensities from the gels are plotted as a percentage of the intensity at time zero for each cell line. (D) Digitonin cell lysates from cells were immunoprecipitated with anti-Ld MAb 64-3-7. Precipitates were then blotted with the antibodies listed. Loading of each sample was normalized on the basis of an equal level of Ld/B7.2 molecules. (E) The Ld/huB7.2-TM construct was introduced into L cells. Surface Ld expression was monitored (MAb 30-5-7) after transduction of the cells with pMIG (vector) or pMIG-mK3. Right panels show anti-tapasin immunoblotting of anti-TAP-1 or anti-Ld precipitates from the indicated cell lines. The Ld/huB7.2-TM construct consists of the luminal domain of Ld (residues 1 to 282) followed by the TM and cytoplasmic tail of human B7.2 (residues 225 to 306).
FIG. 2.
Ld/B7.2 chimeras are sensitive to mK3, and this correlates with their ability to associate with TAP/tapasin. (A) Schematic representation of Ld/B7.2 chimeras constructed with the luminal and TM regions of H-2Ld and cytoplasmic tail of either mouse B7.2 (Ld/mB7.2) or human B7.2 (Ld/huB7.2). A sequence alignment of the cytoplasmic tails of the indicated proteins is shown below the diagram. Lysine residues (K) are shown in bold. Identical or similar residues are shaded. (B) Constructs in panel A were introduced into B6/WT3 cells, followed by transduction with pMIG only (vector) or pMIG-mK3 (mK3). Sorting was used to enrich for GFP+ cells. Cells were incubated for 24 h with 100 U of IFN-γ/ml and then analyzed for expression of wt Ld or the Ld/B7.2 constructs by using MAb 30-5-7. Solid lines, staining of vector-only cells; dashed lines, staining of the mK3-expressing cells. (C) IFN-γ-treated cells were pulse-labeled with [35S]Cys/Met and chased for the indicated times with unlabeled Cys/Met. Ld/huB7.2 molecules were precipitated with MAb 64-3-7. Precipitates were resolved by SDS-PAGE and visualized by autoradiography. Ld/huB7.2 bands are indicated (upper panel). In the lower panel, relative band intensities from the gels are plotted as a percentage of the intensity at time zero for each cell line. (D) Digitonin cell lysates from cells were immunoprecipitated with anti-Ld MAb 64-3-7. Precipitates were then blotted with the antibodies listed. Loading of each sample was normalized on the basis of an equal level of Ld/B7.2 molecules. (E) The Ld/huB7.2-TM construct was introduced into L cells. Surface Ld expression was monitored (MAb 30-5-7) after transduction of the cells with pMIG (vector) or pMIG-mK3. Right panels show anti-tapasin immunoblotting of anti-TAP-1 or anti-Ld precipitates from the indicated cell lines. The Ld/huB7.2-TM construct consists of the luminal domain of Ld (residues 1 to 282) followed by the TM and cytoplasmic tail of human B7.2 (residues 225 to 306).
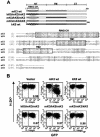
FIG. 3.
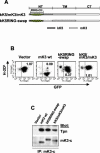
Replacement of the N-terminal (NT), TM, or C-terminal (CT) domain of mK3 with analogous domains of kK5 affects surface downregulation. (A) Schematic description and nomenclature of mK3 chimeras with KSHV kK5 (upper portion). An alignment of mK3 and kK5 sequences is shown in the lower portion. The numbers indicate the first and last amino acids for full-length mK3 or kK5. (B) Each mK3 chimera was introduced into B6/WT3 cells by using the pMIG vector. An analysis of surface Kb expression versus GFP fluorescence is shown. The number in each plot represents the ratio of Kb staining intensities between GFP+ and GFP− populations. Analysis of Db yielded very similar results (data not shown).
FIG. 4.
C-terminal domain of mK3 is required for functional interaction with TAP/tapasin. Each mK3 chimera in Fig. 3A was introduced into β2m−/− cells (H-2b) by using pMIG, followed by sorting to enrich the GFP+ fraction. Following culture with or without 100 U of IFN-γ/ml for 24 h, lysates from the cells were blotted with the indicated antibodies. The GFP signal intensity serves as a loading control. The arrow indicates the position of wt kK5.
FIG. 5.
Information in the final 12 amino acids of mK3 is critical for TAP/tapasin interaction. (A) Schematic depiction of the C-terminal domain (CT) mutants of mK3. NT, N-terminal domain. In the mK3/kK5intCT construct, mK3 residues 140 to 189 were replaced by kK5 residues 147 to 194. (B) Each mutant was introduced into B6/WT3 cells. Flow cytometric analysis of Kb versus GFP fluorescence is shown. The numbers in each plot represent the ratio of Kb staining intensities between GFP+ and GFP− fractions. Analysis of Db expression yielded very similar results (data not shown). (C) The GFP+ cells from each line shown in panel B were enriched by sorting. Lysates from these cells were blotted with the indicated antibodies following culture for 24 h with or without 100 U of IFN-γ/ml. GFP staining levels serve as a loading control as well as an indicator of the effect of IFN-γ on pMIG expression. Bands corresponding to mK3CT190 and mK3CT199 are indicated by arrowheads.
FIG. 6.
Both TAP1 and TAP2 are required for mK3 function. (A) B6/WT3 cells and TAP2-deficient cells (RMA/S) were transduced with mK3 or mK3ΔCT12 (see Fig. 5A) by using pMIG, followed by sorting to enrich for GFP-expressing cells. After culture with or without 100 U of IFN-γ/ml for 24 h, lysates from these cells were immunoblotted with the antibodies listed. GFP staining indicated equivalent expressions of the vector in both lines. The arrow indicates TAP1. (B) Lysates from RMA/S or B6/WT3 (with or without mK3) were immunoprecipitated with the indicated antibodies. Precipitates were blotted as indicated. (C) Kb expression on RMA/S cells with mK3 that were cultured in the presence or absence of the Kb binding peptide OVA (200 μM) (5) with or without 100 U of IFN-γ/ml for 24 h. The numbers represent the geometric mean fluorescence of Kb expression for the GFP+ or GFP− population. (D) RMA/S cells with or without mK3 were pulse-labeled with [35S]Cys/Met and chased for the indicated times. H-2Db immunoprecipitates (MAb 28-14-8) were resolved by SDS-PAGE, and labeled proteins were visualized by autoradiography (upper panel). The lower panel shows the relative band intensities from the gels as a percentage of the intensity at time zero for each cell line.
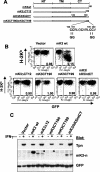
FIG. 7.
Unique information in the N-terminal domain of mK3 not shared with kK5 is involved in its E3 ligase activity. (A) Schematic depiction of two mK3/kK5 chimeras in which different portions of the N-terminal domain of mK3 were replaced by the corresponding portions of kK5. (B) L-Ld cells were transduced with constructs encoding the chimeras or wt mK3, and the GFP+ transductants were enriched by cell sorting. Dk expression (MAb 15-5-5) versus GFP intensity is shown. The numbers indicate the ratio of Dk staining intensities between GFP+ and GFP− populations in each line. Similar profiles were observed with Ld expression (data not shown). (C) Digitonin lysates from the cells in panel B were precipitated with rabbit anti mK3-c serum. Precipitates were then blotted with the antibodies listed. Note that the mK3 blot indicates comparable steady-state levels of both chimeras and wt mK3. (D) Cells were pulse-labeled with [35S]Cys/Met and chased for the indicated times. Lysates were precipitated with a mixture of anti-Ld MAbs 64-3-7 and 30-5-7. Following SDS-PAGE, labeled Ld molecules were visualized by autoradiography (left panel). Relative band intensities from the gels in the left panel are plotted as a percentage of the intensity at time zero for each cell line (right panel). (E) Digitonin lysates from the indicated cell lines were precipitated with MAb 64-3-7. After treatment with or without endo H, precipitates were blotted with either anti-Ub MAb PD41 (upper panel) or anti-Ld (MAb 64-3-7) (lower panel). In the lower panel, R and S indicate endo H-resistant and -sensitive forms of the heavy chain, respectively. Note that ubiquitinated Ld bands can only be detected in the presence of wt mK3 or kK5RING-swap (upper panel), and these bands shift down upon endo H digestion, confirming their endo H-sensitive nature.
FIG. 7.
Unique information in the N-terminal domain of mK3 not shared with kK5 is involved in its E3 ligase activity. (A) Schematic depiction of two mK3/kK5 chimeras in which different portions of the N-terminal domain of mK3 were replaced by the corresponding portions of kK5. (B) L-Ld cells were transduced with constructs encoding the chimeras or wt mK3, and the GFP+ transductants were enriched by cell sorting. Dk expression (MAb 15-5-5) versus GFP intensity is shown. The numbers indicate the ratio of Dk staining intensities between GFP+ and GFP− populations in each line. Similar profiles were observed with Ld expression (data not shown). (C) Digitonin lysates from the cells in panel B were precipitated with rabbit anti mK3-c serum. Precipitates were then blotted with the antibodies listed. Note that the mK3 blot indicates comparable steady-state levels of both chimeras and wt mK3. (D) Cells were pulse-labeled with [35S]Cys/Met and chased for the indicated times. Lysates were precipitated with a mixture of anti-Ld MAbs 64-3-7 and 30-5-7. Following SDS-PAGE, labeled Ld molecules were visualized by autoradiography (left panel). Relative band intensities from the gels in the left panel are plotted as a percentage of the intensity at time zero for each cell line (right panel). (E) Digitonin lysates from the indicated cell lines were precipitated with MAb 64-3-7. After treatment with or without endo H, precipitates were blotted with either anti-Ub MAb PD41 (upper panel) or anti-Ld (MAb 64-3-7) (lower panel). In the lower panel, R and S indicate endo H-resistant and -sensitive forms of the heavy chain, respectively. Note that ubiquitinated Ld bands can only be detected in the presence of wt mK3 or kK5RING-swap (upper panel), and these bands shift down upon endo H digestion, confirming their endo H-sensitive nature.
FIG. 8.
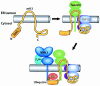
Proposed proximity model depicting how mK3 interacts with host proteins to specifically target the rapid degradation of class I proteins. Upper left, nonconformed mK3. Upper right, mK3 forms a complex with TAP/tapasin in a class I-independent manner. The association of mK3 with TAP/tapasin is primarily mediated through the C-terminal domain of mK3, and this interaction may induce a conformational change within mK3 that not only stabilizes mK3 but also restricts the mobility of its RING-CH domain in the N terminus to achieve the proximity, conformation, and/or orientation required for its E3 ligase function. In this model, tapasin, TAP1, and TAP2 are all essential components of this functional interaction with mK3, although our results suggest that TAP1 and -2 are the direct binding partners of mK3. Bottom, an ER-related E2-conjugating enzyme and/or another necessary factor(s) are recruited by the interactions with the N-terminal region of mK3 (both within and outside the RING-CH domain). These interactions result in the conjugation of Ub to the class I molecules. The Ub conjugation is specific for class I proteins due to a required proximity and/or conformation or orientation imposed by mK3 binding to tapasin/TAP1-tapasin/TAP2 complexes. Thus, substrate specificity is determined by interaction with TAP/tapasin, rather than a specific targeting sequence in the tail of class I.
Similar articles
- Virus subversion of the MHC class I peptide-loading complex.
Lybarger L, Wang X, Harris MR, Virgin HW 4th, Hansen TH. Lybarger L, et al. Immunity. 2003 Jan;18(1):121-30. doi: 10.1016/s1074-7613(02)00509-5. Immunity. 2003. PMID: 12530981 - Requirements for the selective degradation of endoplasmic reticulum-resident major histocompatibility complex class I proteins by the viral immune evasion molecule mK3.
Wang X, Connors R, Harris MR, Hansen TH, Lybarger L. Wang X, et al. J Virol. 2005 Apr;79(7):4099-108. doi: 10.1128/JVI.79.7.4099-4108.2005. J Virol. 2005. PMID: 15767411 Free PMC article. - Downregulation of cell surface receptors by the K3 family of viral and cellular ubiquitin E3 ligases.
Lehner PJ, Hoer S, Dodd R, Duncan LM. Lehner PJ, et al. Immunol Rev. 2005 Oct;207:112-25. doi: 10.1111/j.0105-2896.2005.00314.x. Immunol Rev. 2005. PMID: 16181331 Review. - The role of tapasin in MHC class I antigen assembly.
Androlewicz MJ. Androlewicz MJ. Immunol Res. 1999;20(2):79-88. doi: 10.1007/BF02786464. Immunol Res. 1999. PMID: 10580633 Review.
Cited by
- Stress-regulated targeting of the NKG2D ligand Mult1 by a membrane-associated RING-CH family E3 ligase.
Nice TJ, Deng W, Coscoy L, Raulet DH. Nice TJ, et al. J Immunol. 2010 Nov 1;185(9):5369-76. doi: 10.4049/jimmunol.1000247. Epub 2010 Sep 24. J Immunol. 2010. PMID: 20870941 Free PMC article. - A CD8+ T cell immune evasion protein specific to Epstein-Barr virus and its close relatives in Old World primates.
Hislop AD, Ressing ME, van Leeuwen D, Pudney VA, Horst D, Koppers-Lalic D, Croft NP, Neefjes JJ, Rickinson AB, Wiertz EJ. Hislop AD, et al. J Exp Med. 2007 Aug 6;204(8):1863-73. doi: 10.1084/jem.20070256. Epub 2007 Jul 9. J Exp Med. 2007. PMID: 17620360 Free PMC article. - Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3.
Wang X, Herr RA, Chua WJ, Lybarger L, Wiertz EJ, Hansen TH. Wang X, et al. J Cell Biol. 2007 May 21;177(4):613-24. doi: 10.1083/jcb.200611063. Epub 2007 May 14. J Cell Biol. 2007. PMID: 17502423 Free PMC article. - Applications of major histocompatibility complex class I molecules expressed as single chains.
Primeau T, Myers NB, Yu YY, Lybarger L, Wang X, Truscott SM, Hansen TH, Connolly JM. Primeau T, et al. Immunol Res. 2005;32(1-3):109-21. doi: 10.1385/ir:32:1-3:109. Immunol Res. 2005. PMID: 16106063 Review. - Bovine herpesvirus 1 UL49.5 protein inhibits the transporter associated with antigen processing despite complex formation with glycoprotein M.
Lipińska AD, Koppers-Lalic D, Rychłowski M, Admiraal P, Rijsewijk FA, Bieńkowska-Szewczyk K, Wiertz EJ. Lipińska AD, et al. J Virol. 2006 Jun;80(12):5822-32. doi: 10.1128/JVI.02707-05. J Virol. 2006. PMID: 16731921 Free PMC article.
References
- Androlewicz, M. J., B. Ortmann, P. M. van Endert, T. Spies, and P. Cresswell. 1994. Characteristics of peptide and major histocompatibility complex class I/beta 2-microglobulin binding to the transporters associated with antigen processing (TAP1 and TAP2). Proc. Natl. Acad. Sci. USA 91:12716-12720. - PMC - PubMed
- Boname, J. M., B. D. de Lima, P. J. Lehner, and P. G. Stevenson. 2004. Viral degradation of the MHC class I peptide loading complex. Immunity 20:305-317. - PubMed
- Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627-636. - PubMed
- Cannon, M. J., C. K. Osborn, R. A. Nazaruk, V. Grigoriev, and M. D. Crew. 1999. Diminished recognition of HLA-A2 proteins lacking a cytoplasmic domain (CY) by A2-restricted, EBV-specific CTLs: possible role of the CY in TAP association. Immunogenetics 49:346-350. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI042793/AI/NIAID NIH HHS/United States
- AI46553/AI/NIAID NIH HHS/United States
- R01 AI019687/AI/NIAID NIH HHS/United States
- R01 AI046553/AI/NIAID NIH HHS/United States
- R21 AI042793/AI/NIAID NIH HHS/United States
- AI42793/AI/NIAID NIH HHS/United States
- AI19687/AI/NIAID NIH HHS/United States
- T32AI07063/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous