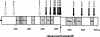Human respiratory coronavirus OC43: genetic stability and neuroinvasion - PubMed (original) (raw)
Comparative Study
Human respiratory coronavirus OC43: genetic stability and neuroinvasion
Julien R St-Jean et al. J Virol. 2004 Aug.
Abstract
The complete genome sequences of the human coronavirus OC43 (HCoV-OC43) laboratory strain from the American Type Culture Collection (ATCC), and a HCoV-OC43 clinical isolate, designated Paris, were obtained. Both genomes are 30,713 nucleotides long, excluding the poly(A) tail, and only differ by 6 nucleotides. These six mutations are scattered throughout the genome and give rise to only two amino acid substitutions: one in the spike protein gene (I958F) and the other in the nucleocapsid protein gene (V81A). Furthermore, the two variants were shown to reach the central nervous system (CNS) after intranasal inoculation in BALB/c mice, demonstrating neuroinvasive properties. Even though the ATCC strain could penetrate the CNS more effectively than the Paris 2001 isolate, these results suggest that intrinsic neuroinvasive properties already existed for the HCoV-OC43 ATCC human respiratory isolate from the 1960s before it was propagated in newborn mouse brains. It also demonstrates that the molecular structure of HCoV-OC43 is very stable in the environment (the two variants were isolated ca. 40 years apart) despite virus shedding and chances of persistence in the host. The genomes of the two HCoV-OC43 variants display 71, 53.1, and 51.2% identity with those of mouse hepatitis virus A59, severe acute respiratory syndrome human coronavirus Tor2 strain (SARS-HCoV Tor2), and human coronavirus 229E (HCoV-229E), respectively. HCoV-OC43 also possesses well-conserved motifs with regard to the genome sequence of the SARS-HCoV Tor2, especially in open reading frame 1b. These results suggest that HCoV-OC43 and SARS-HCoV may share several important functional properties and that HCoV-OC43 may be used as a model to study the biology of SARS-HCoV without the need for level three biological facilities.
Figures
FIG. 1.
Schematic representation of the HCoV-OC43 genome and of the amplification strategy used for sequencing. The HCoV-OC43 genome is 30,713 nt long and comprises nine main ORFs: ORF1a, ORF1b, ns2 (the gene encoding the nonstructural protein 2), HE (hemagglutinin-esterase gene), S (spike gene), ns12.9 (the gene encoding a nonstructural protein of 12.9 kDa), E (small envelope gene), M (membrane gene), and N (nucleocapsid gene). The replicase gene includes both ORF1a and ORF1b. The entire genome was amplified in six fragments in order to be sequenced. Each PCR product was named according to the name of the primers used for the amplification, and the location in the genome is indicated above or below each PCR product. Boxes: open, gene encoding the replicase polyprotein; dotted, genes encoding nonstructural proteins; shaded, genes encoding structural proteins; black, UTRs. GR, GeneRacer.
FIG. 2.
Schematic representation of the polyprotein 1ab putative proteolytic processing and of the main domains found in ORF1ab. The approximate positions of predicted functional domains and protease cleavage sites in ORF1ab are shown, and amino acids positions are also indicated. The white arrows indicate putative cleavage sites recognized either by the PLP1 or the PLP2, whereas black arrows indicate sites recognized by the main protease, 3CLpro. The 15 putative cleavage products generated by the proteolytic processing are named as follows: leader protein, MHV p65-like protein, nsp1 (PL1, X, PL2, and T1), T2, nsp2 (3CLpro), nsp3 (T3), nsp4, nsp5, nsp6, nsp7, nsp9 (RdRp), nsp10 (HEL), nsp11, nsp12, and nsp13 (15). A putative ribosomal −1 frameshift is indicated between ORF1a and ORF1b. Upstream of the frameshift site, the slippery sequence 13334UUUAAAC13340 is found. PL1 and PL2, accessory protease domains; X, conserved domain of unknown function; T1, T2, and T3, membrane-spanning (hydrophobic) domains; 3C, 3CLpro domain; Z, putative zinc finger; HEL, NTPase RNA helicase domain; ND, domain conserved exclusively in nidoviruses. nsp, Nonstructural protein.
FIG. 3.
Neuroinvasive properties of HCoV-OC43 ATCC and HCoV-OC43 Paris variant in BALB/c mice after intranasal inoculation. (A) At 3 dpi, cells positive for viral antigens (arrows) were first observed in the olfactory bulb (OB). No infected cells could be detected in the cortex or other brain structures, illustrating transneuronal spreading of the virus. (B) At 7 dpi, the virus has disseminated to the entire CNS, as illustrated by the presence of immunopositive cells throughout the brain. Magnification (A and B), ×32. (C) Quantification of infectious virus in the brain of each mouse at different times postinfection. Virus titers are presented as logarithmic value of TCID50 per gram of tissue (the limit of detection was 100.5 TCID50/g). Infection by HCoV-OC43 ATCC was detected in one mouse as early as 2 dpi, and gradually more mice became positive. HCoV-OC43 ATCC infectious particles were found between 2 to 8 dpi in mouse brain and led to fatal encephalitis before the end of the experimentation. Antigens of the HCoV-OC43 Paris variant were first revealed in mouse brain at 6 dpi. Infectious particles were detected in some of the brains up to 10 dpi. HCoV-OC43 Paris infectious titers in susceptible animals were similar to those found after HCoV-OC43 ATCC infection, and mice positive for either variant presented all pathological and clinical signs of encephalitis.
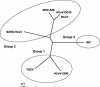
FIG. 4.
Phylogenic unrooted tree regrouping seven coronavirus complete genomes from the three genetic groups. Circles regroup members of each three genetic groups. The 0.1 sliding bar represents the genetic distance between the species (i.e., nucleotide substitution units per studied site). Strains: MHV-A59 (NC_001846); BCoV, bovine coronavirus Quebec strain (AF220295); SARS-HCoV, SARS-HCoV Tor2 strain (AY274119); IBV, IBV Beaudette strain (NC_001451); TGEV (NC_002306); HCoV-229E (NC_002645).
FIG. 5.
Multiple alignments of amino acids of the main proteases of coronaviruses from all three genetic groups. Positions with absolute conservation are shadowed, whereas residues of the putative catalytic dyad, His41 and Cys145, are boxed. Conservation level among group 2 coronaviruses was ca. 46.2%, whereas all strains displayed 26% identity. Strains: OC43, HCoV-OC43 (group 2); BCoV, BCoV Quebec group 2; MHV-A59, MHV group 2; SARS Tor2, SARS-HCoV Tor2 group 2; 229E, HCoV-229E group 1; IBV, IBV group 3.
Comment in
- Genetic variability of human respiratory coronavirus OC43.
Vijgen L, Lemey P, Keyaerts E, Van Ranst M. Vijgen L, et al. J Virol. 2005 Mar;79(5):3223-4; author reply 3224-5. doi: 10.1128/JVI.79.5.3223-3225.2005. J Virol. 2005. PMID: 15709046 Free PMC article. No abstract available.
Similar articles
- Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event.
Vijgen L, Keyaerts E, Moës E, Thoelen I, Wollants E, Lemey P, Vandamme AM, Van Ranst M. Vijgen L, et al. J Virol. 2005 Feb;79(3):1595-604. doi: 10.1128/JVI.79.3.1595-1604.2005. J Virol. 2005. PMID: 15650185 Free PMC article. - Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone.
St-Jean JR, Desforges M, Almazán F, Jacomy H, Enjuanes L, Talbot PJ. St-Jean JR, et al. J Virol. 2006 Apr;80(7):3670-4. doi: 10.1128/JVI.80.7.3670-3674.2006. J Virol. 2006. PMID: 16537637 Free PMC article. - Cleavage of a Neuroinvasive Human Respiratory Virus Spike Glycoprotein by Proprotein Convertases Modulates Neurovirulence and Virus Spread within the Central Nervous System.
Le Coupanec A, Desforges M, Meessen-Pinard M, Dubé M, Day R, Seidah NG, Talbot PJ. Le Coupanec A, et al. PLoS Pathog. 2015 Nov 6;11(11):e1005261. doi: 10.1371/journal.ppat.1005261. eCollection 2015. PLoS Pathog. 2015. PMID: 26545254 Free PMC article. - Human Coronavirus OC43 as a Low-Risk Model to Study COVID-19.
Kim MI, Lee C. Kim MI, et al. Viruses. 2023 Feb 20;15(2):578. doi: 10.3390/v15020578. Viruses. 2023. PMID: 36851792 Free PMC article. Review. - Human coronaviruses: Clinical features and phylogenetic analysis.
Li SW, Lin CW. Li SW, et al. Biomedicine (Taipei). 2013 Mar;3(1):43-50. doi: 10.1016/j.biomed.2012.12.007. Epub 2013 Feb 1. Biomedicine (Taipei). 2013. PMID: 32289002 Free PMC article. Review.
Cited by
- Identification and classification of differentially expressed genes reveal potential molecular signature associated with SARS-CoV-2 infection in lung adenocarcinomal cells.
Soremekun OS, Omolabi KF, Soliman MES. Soremekun OS, et al. Inform Med Unlocked. 2020;20:100384. doi: 10.1016/j.imu.2020.100384. Epub 2020 Jun 23. Inform Med Unlocked. 2020. PMID: 32835074 Free PMC article. - Neurophysiological findings and their prognostic value in critical COVID-19 patients: An observational study.
Niguet JP, Tortuyaux R, Garcia B, Jourdain M, Chaton L, Préau S, Poissy J, Favory R, Nseir S, Mathieu D, Kazali Alidjinou E, Delval A, Derambure P; Lille Intensive Care and Neurophysiology COVID-19 study group. Niguet JP, et al. Clin Neurophysiol. 2021 May;132(5):1009-1017. doi: 10.1016/j.clinph.2021.02.007. Epub 2021 Feb 25. Clin Neurophysiol. 2021. PMID: 33743295 Free PMC article. - Neurotropism of SARS-CoV-2 and its neuropathological alterations: Similarities with other coronaviruses.
Hu J, Jolkkonen J, Zhao C. Hu J, et al. Neurosci Biobehav Rev. 2020 Dec;119:184-193. doi: 10.1016/j.neubiorev.2020.10.012. Epub 2020 Oct 19. Neurosci Biobehav Rev. 2020. PMID: 33091416 Free PMC article. Review. - The acetyl-esterase activity of the hemagglutinin-esterase protein of human coronavirus OC43 strongly enhances the production of infectious virus.
Desforges M, Desjardins J, Zhang C, Talbot PJ. Desforges M, et al. J Virol. 2013 Mar;87(6):3097-107. doi: 10.1128/JVI.02699-12. Epub 2013 Jan 2. J Virol. 2013. PMID: 23283955 Free PMC article. - Molecular Basis of Coronavirus Virulence and Vaccine Development.
Enjuanes L, Zuñiga S, Castaño-Rodriguez C, Gutierrez-Alvarez J, Canton J, Sola I. Enjuanes L, et al. Adv Virus Res. 2016;96:245-286. doi: 10.1016/bs.aivir.2016.08.003. Epub 2016 Aug 30. Adv Virus Res. 2016. PMID: 27712626 Free PMC article. Review.
References
- Addie, D. D., I. A. T. Schaap, L. Nicolson, and O. Jarrett. 2003. Persistence and transmission of natural type I feline coronavirus infection. J. Gen. Virol. 84:2735-2744. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous