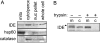Alternative translation initiation generates a novel isoform of insulin-degrading enzyme targeted to mitochondria - PubMed (original) (raw)
Alternative translation initiation generates a novel isoform of insulin-degrading enzyme targeted to mitochondria
Malcolm A Leissring et al. Biochem J. 2004.
Abstract
IDE (insulin-degrading enzyme) is a widely expressed zinc-metallopeptidase that has been shown to regulate both cerebral amyloid beta-peptide and plasma insulin levels in vivo. Genetic linkage and allelic association have been reported between the IDE gene locus and both late-onset Alzheimer's disease and Type II diabetes mellitus, suggesting that altered IDE function may contribute to some cases of these highly prevalent disorders. Despite the potentially great importance of this peptidase to health and disease, many fundamental aspects of IDE biology remain unresolved. Here we identify a previously undescribed mitochondrial isoform of IDE generated by translation at an in-frame initiation codon 123 nucleotides upstream of the canonical translation start site, which results in the addition of a 41-amino-acid N-terminal mitochondrial targeting sequence. Fusion of this sequence to the N-terminus of green fluorescent protein directed this normally cytosolic protein to mitochondria, and full-length IDE constructs containing this sequence were also directed to mitochondria, as revealed by immuno-electron microscopy. Endogenous IDE protein was detected in purified mitochondria, where it was protected from digestion by trypsin and migrated at a size consistent with the predicted removal of the N-terminal targeting sequence upon transport into the mitochondrion. Functionally, we provide evidence that IDE can degrade cleaved mitochondrial targeting sequences. Our results identify new mechanisms regulating the subcellular localization of IDE and suggest previously unrecognized roles for IDE within mitochondria.
Figures
Figure 1. The amino acid and nucleic acid sequences corresponding to the N-terminus of IDE are conserved among mammalian species
(A) The amino acid sequences of human, rat and mouse IDE all feature two alternative in-frame initiating methionines, Met1 and Met42 (bold). The high degree of sequence similarity among the different species implies a functional role for this region. The amino acid mutated in the GK rat (His18→Arg) is highlighted in black. The epitope for the antibody IDE-5 is underlined. The arrow indicates the predicted cleavage site of the Met1-IDE isoform by MPP. (B) The mRNA sequence flanking the second initiation codon (encoding Met42) is closer to the Kozak consensus sequence than that flanking the first initiation codon (encoding Met1). The initiation codons are underlined, and nucleic acids matching the Kozak consensus sequence are shown in bold. Purines (A or G) are denoted by P.
Figure 2. The N-terminus of the Met1-IDE isoform contains a mitochondrial targeting sequence
(A) Design of EGFP fusion constructs used to test for potential sorting sequences within the N-terminus of Met1-IDE or Met42-IDE. EGFP alone served as a control for cytosolic localization. (B) Confocal microscopy showing the subcellular localization of EGFP alone (upper left panel), Met42-IDENTF::EGFP (upper middle panel) and Met1-IDENTF::EGFP (upper right panel). The lower panels shows a cell co-transfected with Met1-IDENTF::EGFP and the mitochondrion-directed DsRed fluorescent protein.
Figure 3. The full-length Met1-IDE isoform is expressed in a cell fraction enriched in mitochondria
(A) Design of full-length, HA-tagged IDE expression constructs designed to exclusively express the Met1-IDE and Met42-IDE isoforms. Note that the first initiation codon is deleted in the Met42-HA–IDE construct, while Met42 is mutated to Ala within the Met1-HA–IDE and Koz-Met1-HA–IDE constructs. The wtHA–IDE construct contains both initiation codons. (B) Western blots showing expression of the different HA-tagged constructs in fractions (10 μg/well) generated by differential centrifugation. Labels to the left of (A) apply also to (B). P2, post-nuclear 3000 g pellet; P3, 10000 g pellet; P4, 100000 g pellet; S1, 100000 g supernatant fraction. Note that the Met42-HA–IDE construct is expressed predominantly in the S1 fraction, which contains exclusively soluble (cytosolic) proteins, while the Koz-Met1-HA–IDE construct is expressed almost exclusively in the P2 pellet, which is highly enriched in mitochondria (see D). (C) The Met1-HA–IDE construct (lacking a Kozak consensus sequence) is expressed at low levels in the P2 fraction. The panel shows an overexposed anti-HA Western blot (30 μg/well) of fractionated cells expressing Met1-HA–IDE compared with empty vector. The Met1-HA–IDE isoform is increased exclusively in the P2 fraction (arrow). Note the presence of non-specific bands (*) of equal intensity in the two P2 fractions. (D) The relative distribution of different subcellular compartments in our fractionation method is exemplified by fractions from the Koz-Met1-HA–IDE cell fractions subjected to Western analysis with several subcellular markers: hsp60 (mitochondrial matrix protein; enriched in P2), catalase (peroxisomal marker; enriched in P3) and α-syntaxin 13 (α-synt 13; endosomal marker; enriched in P4).
Figure 4. Endogenous IDE is present in purified mitochondria
(A) Detection of IDE in purified mitochondria. Note that mitochondrial IDE migrates at a similar size as cytosolic/peroxisomal IDE. (B) Mitochondrial (m) but not cytosolic (c) IDE is protected from digestion with trypsin. The arrowhead indicates an IDE species of a size consistent with that predicted for uncleaved Met1-IDE (i.e. still containing the mitochondrial targeting sequence). Note the absence of the latter species in the cytosolic IDE and in the mitochondrial IDE treated with trypsin.
Figure 5. Detection of the mitochondrial isoform of IDE by immuno-electron microscopy
Electron micrographs of CHO cells expressing wt-HA–IDE (A) or Koz-Met1-HA–IDE (B) and labelled with gold-coupled anti-HA antibodies. (A) Wild-type IDE is present in cytosol (c), as expected, but also in mitochondria (m), with a much weaker signal in the nucleus (n). (B) The Met1-HA–IDE isoform is expressed exclusively in mitochondria. Scale bars=200 nm.
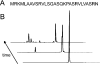
Figure 6. IDE efficiently degrades a prototypical mitochondrial targeting sequence
(A) Amino acid sequence of the cleaved leader peptide of the E1α subunit of human pyruvate dehydrogenase. (B) HPLC elution profiles illustrating that the peptide in (A) is hydrolysed by IDE at multiple sites in a time-dependent fashion.
Similar articles
- Alternative splicing of human insulin-degrading enzyme yields a novel isoform with a decreased ability to degrade insulin and amyloid beta-protein.
Farris W, Leissring MA, Hemming ML, Chang AY, Selkoe DJ. Farris W, et al. Biochemistry. 2005 May 3;44(17):6513-25. doi: 10.1021/bi0476578. Biochemistry. 2005. PMID: 15850385 - Insulin-degrading enzyme.
Authier F, Posner BI, Bergeron JJ. Authier F, et al. Clin Invest Med. 1996 Jun;19(3):149-60. Clin Invest Med. 1996. PMID: 8724818 Review. - Partial loss-of-function mutations in insulin-degrading enzyme that induce diabetes also impair degradation of amyloid beta-protein.
Farris W, Mansourian S, Leissring MA, Eckman EA, Bertram L, Eckman CB, Tanzi RE, Selkoe DJ. Farris W, et al. Am J Pathol. 2004 Apr;164(4):1425-34. doi: 10.1016/s0002-9440(10)63229-4. Am J Pathol. 2004. PMID: 15039230 Free PMC article. - Insulin-degrading enzyme is differentially expressed and developmentally regulated in various rat tissues.
Kuo WL, Montag AG, Rosner MR. Kuo WL, et al. Endocrinology. 1993 Feb;132(2):604-11. doi: 10.1210/endo.132.2.7678795. Endocrinology. 1993. PMID: 7678795 - Multiple functions of insulin-degrading enzyme: a metabolic crosslight?
Tundo GR, Sbardella D, Ciaccio C, Grasso G, Gioia M, Coletta A, Polticelli F, Di Pierro D, Milardi D, Van Endert P, Marini S, Coletta M. Tundo GR, et al. Crit Rev Biochem Mol Biol. 2017 Oct;52(5):554-582. doi: 10.1080/10409238.2017.1337707. Epub 2017 Jun 21. Crit Rev Biochem Mol Biol. 2017. PMID: 28635330 Review.
Cited by
- The Insulin-Degrading Enzyme from Structure to Allosteric Modulation: New Perspectives for Drug Design.
Tundo GR, Grasso G, Persico M, Tkachuk O, Bellia F, Bocedi A, Marini S, Parravano M, Graziani G, Fattorusso C, Sbardella D. Tundo GR, et al. Biomolecules. 2023 Oct 7;13(10):1492. doi: 10.3390/biom13101492. Biomolecules. 2023. PMID: 37892174 Free PMC article. Review. - The amino terminus extension in the long dipeptidyl peptidase 9 isoform contains a nuclear localization signal targeting the active peptidase to the nucleus.
Justa-Schuch D, Möller U, Geiss-Friedlander R. Justa-Schuch D, et al. Cell Mol Life Sci. 2014 Sep;71(18):3611-26. doi: 10.1007/s00018-014-1591-6. Epub 2014 Feb 23. Cell Mol Life Sci. 2014. PMID: 24562348 Free PMC article. - Modulation of Insulin Sensitivity by Insulin-Degrading Enzyme.
González-Casimiro CM, Merino B, Casanueva-Álvarez E, Postigo-Casado T, Cámara-Torres P, Fernández-Díaz CM, Leissring MA, Cózar-Castellano I, Perdomo G. González-Casimiro CM, et al. Biomedicines. 2021 Jan 17;9(1):86. doi: 10.3390/biomedicines9010086. Biomedicines. 2021. PMID: 33477364 Free PMC article. Review. - Immunohistochemical evidence of ubiquitous distribution of the metalloendoprotease insulin-degrading enzyme (IDE; insulysin) in human non-malignant tissues and tumor cell lines.
Weirich G, Mengele K, Yfanti C, Gkazepis A, Hellmann D, Welk A, Giersig C, Kuo WL, Rosner MR, Tang WJ, Schmitt M. Weirich G, et al. Biol Chem. 2008 Nov;389(11):1441-5. doi: 10.1515/BC.2008.157. Biol Chem. 2008. PMID: 18783335 Free PMC article. - Synthesis of two SAPAP3 isoforms from a single mRNA is mediated via alternative translational initiation.
Chua JJ, Schob C, Rehbein M, Gkogkas CG, Richter D, Kindler S. Chua JJ, et al. Sci Rep. 2012;2:484. doi: 10.1038/srep00484. Epub 2012 Jul 2. Sci Rep. 2012. PMID: 22761992 Free PMC article.
References
- Duckworth W. C., Bennett R. G., Hamel F. G. Insulin degradation: progress and potential. Endocr. Rev. 1998;19:608–624. - PubMed
- Qiu W. Q., Ye Z., Kholodenko D., Seubert P., Selkoe D. J. Degradation of amyloid β-protein by a metalloprotease secreted by microglia and other neural and non-neural cells. J. Biol. Chem. 1997;272:6641–6646. - PubMed
- Seta K. A., Roth R. A. Overexpression of insulin degrading enzyme: cellular localization and effects on insulin signaling. Biochem. Biophys. Res. Commun. 1997;231:167–171. - PubMed
- Chesneau V., Perlman R. K., Li W., Keller G. A., Rosner M. R. Insulin-degrading enzyme does not require peroxisomal localization for insulin degradation. Endocrinology. 1997;138:3444–3451. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases