AdoMet radical proteins--from structure to evolution--alignment of divergent protein sequences reveals strong secondary structure element conservation - PubMed (original) (raw)
AdoMet radical proteins--from structure to evolution--alignment of divergent protein sequences reveals strong secondary structure element conservation
Yvain Nicolet et al. Nucleic Acids Res. 2004.
Abstract
Eighteen subclasses of S-adenosyl-l-methionine (AdoMet) radical proteins have been aligned in the first bioinformatics study of the AdoMet radical superfamily to utilize crystallographic information. The recently resolved X-ray structure of biotin synthase (BioB) was used to guide the multiple sequence alignment, and the recently resolved X-ray structure of coproporphyrinogen III oxidase (HemN) was used as the control. Despite the low 9% sequence identity between BioB and HemN, the multiple sequence alignment correctly predicted all but one of the core helices in HemN, and correctly predicted the residues in the enzyme active site. This alignment further suggests that the AdoMet radical proteins may have evolved from half-barrel structures (alphabeta)4 to three-quarter-barrel structures (alphabeta)6 to full-barrel structures (alphabeta)8. It predicts that anaerobic ribonucleotide reductase (RNR) activase, an ancient enzyme that, it has been suggested, serves as a link between the RNA and DNA worlds, will have a half-barrel structure, whereas the three-quarter barrel, exemplified by HemN, will be the most common architecture for AdoMet radical enzymes, and fewer members of the superfamily will join BioB in using a complete (alphabeta)8 TIM-barrel fold to perform radical chemistry. These differences in barrel architecture also explain how AdoMet radical enzymes can act on substrates that range in size from 10 atoms to 608 residue proteins.
Figures
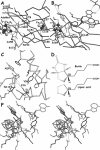
Figure 1
Cα trace of the _Ec_BioB structure. Conserved glycine and/or proline residues within the BioB subclass are represented by a sphere at their Cα position. Strands are depicted in red, helices in blue and loops in black. The Fe4S4 binding loop is depicted in purple.
Figure 2
Multiple sequence alignment containing 18 different AdoMet radical subclasses. For clarity, only one member per subclass is presented here. Each individual selected sequence can be related to the other members of its subclass using standard sequence alignment programs. The alignment corresponds only to structurally homologous sequences in order to link AdoMet radical protein sequences to the _Ec_BioB structure. The numbers in red boxes correspond to the number of omitted residues that were not found to be structurally homologous to the _Ec_BioB structure. The light green boxes correspond to the highly conserved blocks between the different AdoMet radical protein subclasses. The black dashes indicate gaps at these positions. The colored boxes are derived from CLUSTAL (16) and correspond to conserved hydrophobic or aromatic residues (blue), conserved acidic residues (purple), conserved serines or threonines (green), conserved cysteines (pink), conserved lysines or arginines (light red), prolines (yellow) and glycines (orange). Only the parts of the sequences corresponding to the TIM-barrel-like domain are presented here. The secondary structure elements deduced from the _Ec_BioB structure are shown at the top of the alignment, and the ones from _Ec_HemN at the bottom. The parts of the _Ec_HemN sequence represented in gray correspond to fragments that we either misassigned or did not assign. The sequences presented here correspond to E.coli biotin synthase (GI:16128743), the M.jannaschii protein of unknown function (sp|Q58195|), the putative E.coli thiamin synthase (sp|P30140|), the D.vulgaris protein of unknown function (contig 1529 obtained from The Institute for Genomic Research website at
), the M.mazei PylB protein (tr|Q8PWY2|), the M.jannaschii CofG and CofH subunits of the 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase (sp|Q57888| and sp|Q58826|), the E.coli lipoate synthase (sp|P25845|, the E. coli MiaB protein (GI:1786882), the E.coli MoaA protein (sp|P30745|), the A.vinelandii NifB protein (sp|P11067|), the E.coli HemN protein (sp|P32131|), the Clostridium subterminal lysine 2,3-aminomutase (GI:5410603), the E.coli pyruvate formate-lyase activase (sp|P09374|), the T.aromatica benzylsuccinate synthase activase (tr|O87941|), the E.coli class III RNR activase (GI:16132059), the Pseudomonas aeruginosa NirJ protein (tr|P95416|) and the Bacillus subtilis spore photoproduct lyase (GI:16078457).
Figure 3
(A and B) Views of the _Ec_BioB and _Ec_HemN structures, respectively. Helices are depicted in blue, strands in red, and loops in black. The numbers indicate the strand number from the N-terminal extremity of the TIM barrel. The zones interacting with AdoMet are depicted in orange. The secondary structural elements that are different in the two structures are depicted in semi-transparent dark blue. (C and D) Views of protein:AdoMet (green) interactions in _Ec_BioB. (E and F) Same views as (C and D) for _Ec_HemN. The color code for secondary structures is the same as in (A).
Figure 4
(A–C) A view of the active sites of _Ec_BioB, _Ec_HemN and _Dd_NDPk, respectively, based on the superposition of their ribose moiety. AdoMet and ADP are depicted in green. (D) Comparison of the structures of biotin and lipoic acid. (E) Stereoview of the superposition of _Ec_BioB, _Ec_HemN and _Dd_NDPk in green, orange and blue, respectively, in the same orientation as in (A–C).
Figure 5
(A) A view of the _Ec_BioB structure showing the more open side with shorter strands 7 and 8, and helices. The loop proposed to ‘close the door’ of the active site upon substrate binding is depicted in red. (B) A view of _Ec_HemN in the same orientation as _Ec_BioB in (A) showing the open side of the active site cavity. The C-terminal region is depicted in purple. (C) Stereoview of the superposition of the strands of the TIM barrel from _Ec_BioB (green) and the equivalent the three-quarter barrel from _Ec_HemN (light purple). The additional secondary structural elements that complement the three-quarter barrel structure in _Ec_HemN are depicted in red.
Similar articles
- Structure of an archaeal TYW1, the enzyme catalyzing the second step of wye-base biosynthesis.
Goto-Ito S, Ishii R, Ito T, Shibata R, Fusatomi E, Sekine SI, Bessho Y, Yokoyama S. Goto-Ito S, et al. Acta Crystallogr D Biol Crystallogr. 2007 Oct;63(Pt 10):1059-68. doi: 10.1107/S0907444907040668. Epub 2007 Sep 19. Acta Crystallogr D Biol Crystallogr. 2007. PMID: 17881823 - Ribonucleotide reductases: divergent evolution of an ancient enzyme.
Torrents E, Aloy P, Gibert I, Rodríguez-Trelles F. Torrents E, et al. J Mol Evol. 2002 Aug;55(2):138-52. doi: 10.1007/s00239-002-2311-7. J Mol Evol. 2002. PMID: 12107591 - Radical S-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-L-methionines.
Layer G, Grage K, Teschner T, Schünemann V, Breckau D, Masoumi A, Jahn M, Heathcote P, Trautwein AX, Jahn D. Layer G, et al. J Biol Chem. 2005 Aug 12;280(32):29038-46. doi: 10.1074/jbc.M501275200. Epub 2005 Jun 20. J Biol Chem. 2005. PMID: 15967800 - S-adenosylmethionine radical enzymes.
Marsh EN, Patwardhan A, Huhta MS. Marsh EN, et al. Bioorg Chem. 2004 Oct;32(5):326-40. doi: 10.1016/j.bioorg.2004.06.001. Bioorg Chem. 2004. PMID: 15381399 Review. - Structure and function of radical SAM enzymes.
Layer G, Heinz DW, Jahn D, Schubert WD. Layer G, et al. Curr Opin Chem Biol. 2004 Oct;8(5):468-76. doi: 10.1016/j.cbpa.2004.08.001. Curr Opin Chem Biol. 2004. PMID: 15450488 Review.
Cited by
- Natural history of S-adenosylmethionine-binding proteins.
Kozbial PZ, Mushegian AR. Kozbial PZ, et al. BMC Struct Biol. 2005 Oct 14;5:19. doi: 10.1186/1472-6807-5-19. BMC Struct Biol. 2005. PMID: 16225687 Free PMC article. - Complex biotransformations catalyzed by radical S-adenosylmethionine enzymes.
Zhang Q, Liu W. Zhang Q, et al. J Biol Chem. 2011 Sep 2;286(35):30245-30252. doi: 10.1074/jbc.R111.272690. Epub 2011 Jul 19. J Biol Chem. 2011. PMID: 21771780 Free PMC article. Review. - Post-translational modification of ribosomal proteins: structural and functional characterization of RimO from Thermotoga maritima, a radical S-adenosylmethionine methylthiotransferase.
Arragain S, Garcia-Serres R, Blondin G, Douki T, Clemancey M, Latour JM, Forouhar F, Neely H, Montelione GT, Hunt JF, Mulliez E, Fontecave M, Atta M. Arragain S, et al. J Biol Chem. 2010 Feb 19;285(8):5792-801. doi: 10.1074/jbc.M109.065516. Epub 2009 Dec 9. J Biol Chem. 2010. PMID: 20007320 Free PMC article. - The B12-independent glycerol dehydratase activating enzyme from Clostridium butyricum cleaves SAM to produce 5'-deoxyadenosine and not 5'-deoxy-5'-(methylthio)adenosine.
Walls WG, Moody JD, McDaniel EC, Villanueva M, Shepard EM, Broderick WE, Broderick JB. Walls WG, et al. J Inorg Biochem. 2022 Feb;227:111662. doi: 10.1016/j.jinorgbio.2021.111662. Epub 2021 Nov 12. J Inorg Biochem. 2022. PMID: 34847521 Free PMC article. - Radical-mediated enzymatic methylation: a tale of two SAMS.
Zhang Q, van der Donk WA, Liu W. Zhang Q, et al. Acc Chem Res. 2012 Apr 17;45(4):555-64. doi: 10.1021/ar200202c. Epub 2011 Nov 18. Acc Chem Res. 2012. PMID: 22097883 Free PMC article. Review.
References
- Sofia H.J., Chen,G., Hetzler,B.G., Reyes-Spindola,J.F. and Miller,N.E. (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res., 29, 1097–1106. - PMC - PubMed
- Frey P.A. and Booker,S.J. (2001) Radical mechanisms of S-adenosylmethionine-dependent enzymes. Adv. Protein Chem., 58, 1–45. - PubMed
- Hewitson K.S., Baldwin,J.E., Shaw,N.M. and Roach,P.L. (2000) Mutagenesis of the proposed iron–sulfur cluster binding ligands in Escherichia coli biotin synthase. FEBS Lett., 466, 372–376. - PubMed
- Hewitson K.S., Ollagnier-de Choudens,S., Sanakis,Y., Shaw,N.M., Baldwin,J.E., Munck,E., Roach,P.L. and Fontecave,M. (2002) The iron–sulfur center of biotin synthase: site-directed mutants. J. Biol. Inorg. Chem., 7, 83–93. - PubMed
- Layer G., Verfurth,K., Mahlitz,E. and Jahn,D. (2002) Oxygen-independent coproporphyrinogen-III oxidase HemN from Escherichia coli. J. Biol. Chem., 277, 34136–34142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases