CisModule: de novo discovery of cis-regulatory modules by hierarchical mixture modeling - PubMed (original) (raw)
CisModule: de novo discovery of cis-regulatory modules by hierarchical mixture modeling
Qing Zhou et al. Proc Natl Acad Sci U S A. 2004.
Abstract
The regulatory information for a eukaryotic gene is encoded in cis-regulatory modules. The binding sites for a set of interacting transcription factors have the tendency to colocalize to the same modules. Current de novo motif discovery methods do not take advantage of this knowledge. We propose a hierarchical mixture approach to model the cis-regulatory module structure. Based on the model, a new de novo motif-module discovery algorithm, CisModule, is developed for the Bayesian inference of module locations and within-module motif sites. Dynamic programming-like recursions are developed to reduce the computational complexity from exponential to linear in sequence length. By using both simulated and real data sets, we demonstrate that CisModule is not only accurate in predicting modules but also more sensitive in detecting motif patterns and binding sites than standard motif discovery methods are.
Figures
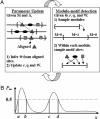
Fig. 1.
Specification of the HMx model. (A) Unaligned motif sites (triangles indexed by 1, 2,... ,5).(B) The aligned motif sites can be represented by a product multinomial model or equivalently by a PWM. Each binding site is regarded as a realization of a sequence of independent random variables X_1X2...Xw, where each Xi (i = 1,..., w) follows a multinomial distribution over the four letters {A,C,G,T} with probabilities θ_i = [θ_i_(A), θ_i_(C), θ_i_(G), θ_i_(T)]. The whole motif is thus specified by a set of multinomial probabilities Θ = [θ1, θ2,..., θ_w_]. (C) The cis-regulatory regions of coregulated genes are enriched for modules (the regions in the brackets). Each module is a sequence segment _x_1_x_2...xl in which several types of motifs (A, B, and C), each with its own product multinomial parameter (Θk), can occur. The rates of the occurrence of modules and their motif sites are denoted by r and qk (k = 1,..., K), respectively.
Fig. 2.
Algorithm for model fitting and motif-module identification. (A) Iterative sampling procedure. In parameter update (Left), we are given the locations of modules and motif sites. Therefore, we align the motif sites of the same type to update the PWM of that motif. In module and motif detection (Right), we use stochastic recursions (see Appendix B and text) to sample the locations of modules and motif sites, conditional on the updated parameter values. (B) The use of sampled module indicators for module identification. For each position i in the sequences, compute Pm(i) = the proportion of times during iterative sampling when position i is within a sampled module. The positions with Pm(i) > 0.5 (e.g., the regions [a,b] and [c,d]) are our predicted modules. See Fig. 3_A_ for further discussion.
Fig. 3.
Module prediction in the Drosophila data set. (A) Marginal posterior module probability (Pm) plots for example sequences in the three data sets of Drosophila homotypic modules. Pm is the probability of being sampled as within modules and it is plotted as a function of the position in the sequences (the solid curves). The horizontal broken lines correspond to Pm = 0.5, and the sequence bases with Pm > 0.5 are our predicted modules. The vertical lines are the motif sites predicted by CisModule. (B) Top site density of S(x) vs. cutoff value x. The broken vertical line at x = 0.5 corresponds to that of Pm = 0.5 in A.
Similar articles
- HeliCis: a DNA motif discovery tool for colocalized motif pairs with periodic spacing.
Larsson E, Lindahl P, Mostad P. Larsson E, et al. BMC Bioinformatics. 2007 Oct 28;8:418. doi: 10.1186/1471-2105-8-418. BMC Bioinformatics. 2007. PMID: 17963524 Free PMC article. - Cross-species de novo identification of cis-regulatory modules with GibbsModule: application to gene regulation in embryonic stem cells.
Xie D, Cai J, Chia NY, Ng HH, Zhong S. Xie D, et al. Genome Res. 2008 Aug;18(8):1325-35. doi: 10.1101/gr.072769.107. Epub 2008 May 15. Genome Res. 2008. PMID: 18490265 Free PMC article. - Inferring intra-motif dependencies of DNA binding sites from ChIP-seq data.
Eggeling R, Roos T, Myllymäki P, Grosse I. Eggeling R, et al. BMC Bioinformatics. 2015 Nov 9;16:375. doi: 10.1186/s12859-015-0797-4. BMC Bioinformatics. 2015. PMID: 26552868 Free PMC article. - Challenges for modeling global gene regulatory networks during development: insights from Drosophila.
Wilczynski B, Furlong EE. Wilczynski B, et al. Dev Biol. 2010 Apr 15;340(2):161-9. doi: 10.1016/j.ydbio.2009.10.032. Epub 2009 Oct 27. Dev Biol. 2010. PMID: 19874814 Review. - Eukaryotic transcription factor binding sites--modeling and integrative search methods.
Hannenhalli S. Hannenhalli S. Bioinformatics. 2008 Jun 1;24(11):1325-31. doi: 10.1093/bioinformatics/btn198. Epub 2008 Apr 21. Bioinformatics. 2008. PMID: 18426806 Review.
Cited by
- Integrating phosphorylation network with transcriptional network reveals novel functional relationships.
Wang L, Hou L, Qian M, Deng M. Wang L, et al. PLoS One. 2012;7(3):e33160. doi: 10.1371/journal.pone.0033160. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22432002 Free PMC article. - Markov chain-based promoter structure modeling for tissue-specific expression pattern prediction.
Vandenbon A, Miyamoto Y, Takimoto N, Kusakabe T, Nakai K. Vandenbon A, et al. DNA Res. 2008 Feb 29;15(1):3-11. doi: 10.1093/dnares/dsm034. Epub 2008 Feb 7. DNA Res. 2008. PMID: 18258700 Free PMC article. - Clustered ChIP-Seq-defined transcription factor binding sites and histone modifications map distinct classes of regulatory elements.
Rye M, Sætrom P, Håndstad T, Drabløs F. Rye M, et al. BMC Biol. 2011 Nov 24;9:80. doi: 10.1186/1741-7007-9-80. BMC Biol. 2011. PMID: 22115494 Free PMC article. - Statistical significance of cis-regulatory modules.
Schones DE, Smith AD, Zhang MQ. Schones DE, et al. BMC Bioinformatics. 2007 Jan 22;8:19. doi: 10.1186/1471-2105-8-19. BMC Bioinformatics. 2007. PMID: 17241466 Free PMC article. - Practical strategies for discovering regulatory DNA sequence motifs.
MacIsaac KD, Fraenkel E. MacIsaac KD, et al. PLoS Comput Biol. 2006 Apr;2(4):e36. doi: 10.1371/journal.pcbi.0020036. PLoS Comput Biol. 2006. PMID: 16683017 Free PMC article. No abstract available.
References
- Hertz, G. Z. & Stormo, G. D. (1999) Bioinformatics 15, 563-577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases