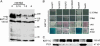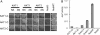K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions - PubMed (original) (raw)
. 2004 Aug 17;101(33):12242-7.
doi: 10.1073/pnas.0404467101. Epub 2004 Aug 6.
Mohamed El-Bakkoury, Tanja Hamacher, Corinna Cappellaro, Cristina Vilarino, Carola Fleischer, Heinz Ellerbrok, Richard Kamuzinzi, Valérie Ledent, Damien Blaudez, Dale Sanders, Jose L Revuelta, Eckhard Boles, Bruno André, Wolf B Frommer
Affiliations
- PMID: 15299147
- PMCID: PMC514463
- DOI: 10.1073/pnas.0404467101
K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions
Petr Obrdlik et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Organization of proteins into complexes is crucial for many cellular functions. However, most proteomic approaches primarily detect protein interactions for soluble proteins but are less suitable for membrane-associated complexes. Here we describe a mating-based split ubiquitin system (mbSUS) for systematic identification of interactions between membrane proteins as well as between membrane and soluble proteins. mbSUS allows in vivo cloning of PCR products into a vector set, detection of interactions via mating, regulated expression of baits, and improved selection of interacting proteins. Cloning is simplified by introduction of lambda attachment sites for GATEWAY. Homo- and heteromeric interactions between Arabidopsis K(+) channels KAT1, AKT1, and AKT2 were identified. Tests with deletion mutants demonstrate that the C terminus of KAT1 and AKT1 is necessary for physical assembly of complexes. Screening of a sorted collection of 84 plant proteins with K(+) channels as bait revealed differences in oligomerization between KAT1, AKT1, and AtKC1, and allowed detection of putative interacting partners of KAT1 and AtKC1. These results show that mbSUS is suited for systematic analysis of membrane protein interactions.
Figures
Fig. 1.
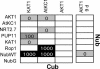
Expression cassettes of pSUgate vectors. pNXgate and pXNgate vectors are suitable for “NubG-X” and “X-NubG” fusions of prey polypeptides “X,” and pMetYCgate is suited for “Y-CubPLV” fusions of bait peptides “Y.” B1-KanMX-B2 is identical in all vectors. Promoters are red, terminators are purple; NubG, Cub, and the linkers are shown on the right. Marked restriction sites are used to produce linear vectors for in vivo cloning. “ATG” and “stop” mark the start and the stop codons in the expression cassettes, respectively. AmpR refers to ampicillin resistance cassette. 2μ refers to a high-copy, whereas CEN/ARS refers to a low-copy, yeast origin of replication. The scale gives length in base pairs.
Fig. 2.
Interaction of KAT1-CubPLV with different NubG-X fusions (A, adenine; H, histidine; N, NubG; C, CubPLV). (A) Expression of met25-KAT1-CubPLV is regulated by Met. Diploid cells carrying KAT1-CubPLV and pNXgate plasmid were grown on synthetic complete -TLU (tryptophan, leucine, uracil) media and different concentrations of Met. Western analysis of cell extracts with anti-PLV antibody was performed; lane d indicates diploids with empty pMetYCgate and pNXgate. Size of proteins is given in kDa (130 kDa corresponds to KAT1-CubPLV fusion; other polypeptides are unspecific because they are present in control). (B) Growth on minimal medium (lacking amino acids) reporting KAT1-CubPLV interactions. Interactions were selected on media with and without 0.15 mM Met. +AH shows the growth of diploids on nonselective minimal medium. X-Gal shows detection of lacZ activity with 5-bromo-4-chloro-3-indolyl β-
d
-galactosidase (X-Gal) as a substrate. Soluble NubWT and NubG were positive and negative controls, respectively. (C) Western analysis of interaction-dependent PLV cleavage with anti-PLV antibody. Extracts were prepared from diploid cells grown on minimal medium +AH. Order of lanes corresponds to B.
Fig. 3.
Screening reveals oligomerization of plant K+ channels KAT1 and AKT1. (A) Flow chart of the applied screening strategy (abbreviations are as in Fig. 2). (B and C) Interaction screen with NubG-X and X-NubG fusion proteins. Replica plates showing diploid cells selected on -TLAH media without Met after 2 (NubG-X) and 9 (X-NubG) days of incubation. NubWT and NubG were positive and negative controls, respectively. (D) Results obtained with CNGC1-CubPLV as bait.
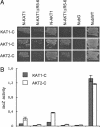
Fig. 4.
Interactions among different KAT1, AKT1, and AKT2 fusions. (A) Diploid cells on minimal media after 4 (KAT1-C and AKT2-C) and 6 (AKT1-C) days. NX are NubG-X fusions, and XN are X-NubG fusions. Abbreviations are as in Fig. 2. (B) Liquid β-galactosidase assay using _o_-nitrophenylglucoside (ONPG) as a substrate. LacZ activity is given in μmol ONPG × min-1 × mg of protein-1. For extracts, cells were grown on -TLAH media without Met. The experiment was repeated twice independently with comparable results.
Fig. 5.
Role of C-terminal R5 and R6 domains in AKT1 and KAT1 oligomerization. (A) Diploid cells on 0.15 mM Met after 3 days (KAT1-C and AKT2-C) and without Met after 5 days (AKT1-C). Abbreviations are as in Fig. 2. (B) Liquid β-galactosidase assays were performed as in Fig. 4_B_. Order of lanes corresponds to A.
Fig. 6.
Interactions of K+ channels detected by screening of a sorted X-NubG collection. Diploids with interacting X-NubG and CubPLV fusions were selected in liquid media with different concentrations of Met. Growth was monitored after 5 days or, for “AKT1 9 d,” after 9 days. The shade of gray and the numbers give maximum in Met concentration (in μM), in which the corresponding diploids were grown.
Similar articles
- AtKC1 is a general modulator of Arabidopsis inward Shaker channel activity.
Jeanguenin L, Alcon C, Duby G, Boeglin M, Chérel I, Gaillard I, Zimmermann S, Sentenac H, Véry AA. Jeanguenin L, et al. Plant J. 2011 Aug;67(4):570-82. doi: 10.1111/j.1365-313X.2011.04617.x. Epub 2011 Jul 11. Plant J. 2011. PMID: 21518051 - Biochemical characterization of the Arabidopsis K+ channels KAT1 and AKT1 expressed or co-expressed in insect cells.
Urbach S, Chérel I, Sentenac H, Gaymard F. Urbach S, et al. Plant J. 2000 Aug;23(4):527-38. doi: 10.1046/j.1365-313x.2000.00828.x. Plant J. 2000. PMID: 10972879 - Multiple genes, tissue specificity, and expression-dependent modulationcontribute to the functional diversity of potassium channels in Arabidopsis thaliana.
Cao Y, Ward JM, Kelly WB, Ichida AM, Gaber RF, Anderson JA, Uozumi N, Schroeder JI, Crawford NM. Cao Y, et al. Plant Physiol. 1995 Nov;109(3):1093-106. doi: 10.1104/pp.109.3.1093. Plant Physiol. 1995. PMID: 8552711 Free PMC article. - K(+) channel profile and electrical properties of Arabidopsis root hairs.
Ivashikina N, Becker D, Ache P, Meyerhoff O, Felle HH, Hedrich R. Ivashikina N, et al. FEBS Lett. 2001 Nov 23;508(3):463-9. doi: 10.1016/s0014-5793(01)03114-3. FEBS Lett. 2001. PMID: 11728473 - Escherichia coli as an expression system for K(+) transport systems from plants.
Uozumi N. Uozumi N. Am J Physiol Cell Physiol. 2001 Sep;281(3):C733-9. doi: 10.1152/ajpcell.2001.281.3.C733. Am J Physiol Cell Physiol. 2001. PMID: 11502550 Review.
Cited by
- OPEN STOMATA 1 phosphorylates CYCLIC NUCLEOTIDE-GATED CHANNELs to trigger Ca2+ signaling for abscisic acid-induced stomatal closure in Arabidopsis.
Yang Y, Tan YQ, Wang X, Li JJ, Du BY, Zhu M, Wang P, Wang YF. Yang Y, et al. Plant Cell. 2024 May 29;36(6):2328-2358. doi: 10.1093/plcell/koae073. Plant Cell. 2024. PMID: 38442317 - ER Membrane Protein Interactions Using the Split-Ubiquitin System (SUS).
Asseck LY, Wallmeroth N, Grefen C. Asseck LY, et al. Methods Mol Biol. 2024;2772:207-219. doi: 10.1007/978-1-0716-3710-4_15. Methods Mol Biol. 2024. PMID: 38411816 - CPK28 is a modulator of purinergic signaling in plant growth and defense.
Sowders JM, Jewell JB, Tanaka K. Sowders JM, et al. Plant J. 2024 May;118(4):1086-1101. doi: 10.1111/tpj.16656. Epub 2024 Feb 3. Plant J. 2024. PMID: 38308597 - The K+ transporter NPF7.3/NRT1.5 and the proton pump AHA2 contribute to K+ transport in Arabidopsis thaliana under K+ and NO3- deficiency.
Sena F, Kunze R. Sena F, et al. Front Plant Sci. 2023 Nov 10;14:1287843. doi: 10.3389/fpls.2023.1287843. eCollection 2023. Front Plant Sci. 2023. PMID: 38046603 Free PMC article. - Comprehensive comparative assessment of the Arabidopsis thaliana MLO2-CALMODULIN2 interaction by various in vitro and in vivo protein-protein interaction assays.
von Bongartz K, Sabelleck B, Baquero Forero A, Kuhn H, Leissing F, Panstruga R. von Bongartz K, et al. Biochem J. 2023 Oct 31;480(20):1615-1638. doi: 10.1042/BCJ20230255. Biochem J. 2023. PMID: 37767715 Free PMC article.
References
- Phizicky, E., Bastiaens, P. I., Zhu, H., Snyder, M. & Fields, S. (2003) Nature 422, 208-215. - PubMed
- Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochrat, P., et al. (2000) Nature 403, 623-627. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous