Direct physical and functional interaction of the NuA4 complex components Yaf9p and Swc4p - PubMed (original) (raw)
Direct physical and functional interaction of the NuA4 complex components Yaf9p and Swc4p
Claudia B Bittner et al. Eukaryot Cell. 2004 Aug.
Abstract
Saccharomyces cerevisiae Yaf9p and the mammalian leukemia-associated protein ENL share a high degree of similarity. To investigate the biological function of Yaf9p, this protein was used to search for interacting proteins in a two-hybrid system. Here, we demonstrate that Yaf9p binds directly to Swc4p, the yeast homolog of the mammalian DNA-methyltransferase-associated protein 1. Yaf9p and Swc4p associate through C-terminal domains, and both proteins coprecipitate in vitro in pull-down experiments and in vivo by immunoprecipitation. In living cells, Swc4p is present in a megadalton protein complex that shows a fractionation behavior in gel filtration similar to that of Esa1p, the histone acetyltransferase of the NuA4 complex. Recruitment of Yaf9p to DNA leads to promoter-specific transcriptional activation that can be inhibited by dominant negative Swc4p lacking the Yaf9p binding domain. Interference with Swc4p function also increases sensitivity to the microtubule toxin benomyl, a trait that corresponds to the known phenotype of a yaf9(-) knockout strain. In summary, the results suggest that Yaf9p and Swc4p form a protein pair that has a role in chromatin modification with possible implications also for the function of their mammalian counterparts.
Figures
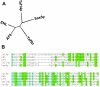
FIG. 1.
Multiple sequence alignment. (A) Dendrogram depicting the relationship of YEATS domain proteins; (B) alignment of the YEATS domain with amino acids given in one-letter codes.
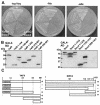
FIG. 2.
Analysis of the Yaf9p/Swc4p interaction in the two-hybrid system. (A) The yeast two-hybrid reporter strain AH109 was transformed with combinations of bait and prey plasmids encoding the following S. cerevisiae proteins: (i) Snf1p as bait and Snf4p as prey (+, positive control for interaction), (ii) full-length Yaf9p as bait and full-length Swc4p as prey, and (iii) human lamin C as bait with Swc4p as prey. Except for the positive control, two independent transformants are shown. Growth was assessed on dropout medium plates lacking tryptophan and leucine (−trp/−leu), histidine (−his), or adenine (−ade). (B) Protein expression of bait (Yaf9p) and prey (Swc4p) derivatives. Various deletion mutants of Yaf9p and Swc4p (numbers indicate respective cDNA positions) were fused either to the DNA BD (GAL4-BD) or the transactivation domain (GAL4-AD) of GAL4 as indicated. Yeast strain AH109 was transformed with the resulting plasmids, and the protein expression was verified by immunoblotting with either an anti-GAL4-DNA BD or an anti-GAL4 AD antibody. (C) Schematic representation of the two-hybrid results obtained with Yaf9p and Swc4p mutants. The bars are drawn to scale. The conserved YEATS domain is indicated by shading. +, growth on double-dropout medium lacking adenine and histidine; −, no growth on selective medium.
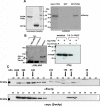
FIG. 3.
In vitro and in vivo interaction between Yaf9p and Swc4p. (A) GST pull-down experiment. Recombinant GST or a fusion of GST with full-length Yaf9p was loaded onto glutathione beads, and contaminating proteins were removed by thorough washing. To ensure proper binding, an aliquot of the charged beads was extracted in SDS sample buffer and analyzed by SDS-gel electrophoresis (left). The prepared beads were incubated with radioactive 35S-labeled Swc4p produced by in vitro transcription and translation. After being washed, the proteins still attached to the beads were eluted, separated by gel electrophoresis, and visualized by autoradiography. As a control, 10% of the input transcription-translation reaction mixture was run alongside the proteins. The positions of molecular mass marker standards are given. (B) Immunoprecipitation (i.p.). Protein extracts from the two-hybrid strain containing GAL4 fusions of Yaf9p (GAL4 BD) and Swc4p (GAL4-AD) were precipitated either with an anti GAL4-DNA BD antibody or with an unspecific control antibody. Proteins in the precipitate were examined for the presence of GAL4 AD-Swc4p by immunoblotting. As a control, an aliquot of protein extract before immunoprecipitation was used (left panel). In a second approach, expression vectors for Flag-tagged Yaf9p were transfected either alone or in combination with a plasmid coding for myc-tagged Swc4p into yeast strain K699, as indicated. Precipitation with an anti-myc antibody pulled down Flag-tagged Yaf9p only if myc-tagged Swc4p was present, indicating a specific interaction between these proteins (right panel). (C) Gel filtration experiment. Native protein extracts from K699 expressing myc-tagged Swc4p were separated on a Superdex 200 sizing column that was calibrated by proteins with a known molecular mass. Outflow fractions were scored for the presence of myc-Swc4p and Esa1p by immunoblotting. The elution positions for blue dextran (molecular mass, 2 MDa) and calibration proteins (see Materials and Methods) are indicated together with the molecular masses of the protein extracts.
FIG. 4.
Functional interaction of Yaf9p and Swc4p. (A) Promoter-specific transactivation capacity of Yaf9p. Full-length Yaf9p fused to the GAL4-DNA BD was transformed into the indicated yeast reporter strains (Yaf9). An empty bait vector (−) or a known positive two-hybrid interaction between the Snf1p and Snf4p proteins (+) served as a negative or positive control, respectively. lacZ and HIS1 reporter gene activation was measured either by determination of β-galactosidase activity or by monitoring growth of the transformants in histidine dropout media. The upstream activation sequences driving reporter expression are indicated for each gene. lacZ units are the means and standard deviation of triplicate samples. For growth assessment, optical density (O.D.) at 600 nm was recorded at the indicated time points. (B) Perturbation of the Yaf9p/Swc4p interaction can interfere with Yaf9p transcriptional activity. A deletion mutant of Swc4p that has lost the C-terminal domain necessary for Yaf9p binding (Swc4Δ) or Swc4p itself (Swc4) was constitutively expressed in the two-hybrid strain PJ69-4A together with GAL4 fusions of either Yaf9p, Gal4p, or Gcn4p as indicated. The expression of the Swc4 proteins was verified by Western blotting (upper left panel). An empty expression vector or a construct producing unrelated GST protein was used as a control. The function of the respective transcriptional activators in the presence ofSwc4Δ or Swc4 was judged by determination of lacZ activity derived from the endogenous _GAL7_-lacZ reporter. Basal activity in the absence of additional proteins was set at 100%. The means and standard deviations of results of triplicate experiments are shown. A schematic representation of the principles of dominant negative and squelching mechanisms is depicted. (C) Perturbation of the Yaf9p/Swc4p interaction can interfere with endogenous Yaf9p function. Yaf9p has been shown to be necessary for resistance against the spindle toxin benomyl (19). Wild-type S. cerevisiae (strain K699, a W303 derivative) was transformed with an empty expression vector (pESC) or with constructs that allowed the overexpression of Swc4Δp and Swc4p under the control of a galactose-inducible promoter. A genomic _yaf9_− mutant in the same genetic background was used as an additional control. Three independent transformants were each plated on medium, with the following results.(Left plate) Galactose medium in the absence of benomyl allows for protein production and controls for a general influence of Swc4p/Swc4Δp on growth. (Center plate) Glucose represses expression of ectopic Swc4p/Swc4Δp, and the addition of benomyl reveals the intrinsic sensitivity of the strains towards spindle stress. (Right plate) Galactose induction of Swc4p/Swc4Δp in the presence of benomyl increases the sensitivity of the respective transformants to spindle stress compared to that of cells treated with vector only.
Similar articles
- Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex.
Nourani A, Doyon Y, Utley RT, Allard S, Lane WS, Côté J. Nourani A, et al. Mol Cell Biol. 2001 Nov;21(22):7629-40. doi: 10.1128/MCB.21.22.7629-7640.2001. Mol Cell Biol. 2001. PMID: 11604499 Free PMC article. - The nuclear actin-related protein of Saccharomyces cerevisiae, Arp4, directly interacts with the histone acetyltransferase Esa1p.
Steinboeck F, Bogusch A, Kaufmann A, Heidenreich E. Steinboeck F, et al. J Biochem. 2007 May;141(5):661-8. doi: 10.1093/jb/mvm083. Epub 2007 Mar 29. J Biochem. 2007. PMID: 17395615 - NuA4 and SWR1-C: two chromatin-modifying complexes with overlapping functions and components.
Lu PY, Lévesque N, Kobor MS. Lu PY, et al. Biochem Cell Biol. 2009 Oct;87(5):799-815. doi: 10.1139/O09-062. Biochem Cell Biol. 2009. PMID: 19898529 Review. - Structure of histone acetyltransferases.
Marmorstein R. Marmorstein R. J Mol Biol. 2001 Aug 17;311(3):433-44. doi: 10.1006/jmbi.2001.4859. J Mol Biol. 2001. PMID: 11492997 Review.
Cited by
- CaSWC4 regulates the immunity-thermotolerance tradeoff by recruiting CabZIP63/CaWRKY40 to target genes and activating chromatin in pepper.
Cai W, Yang S, Wu R, Zheng Y, He S, Shen L, Guan D, He S. Cai W, et al. PLoS Genet. 2022 Feb 28;18(2):e1010023. doi: 10.1371/journal.pgen.1010023. eCollection 2022 Feb. PLoS Genet. 2022. PMID: 35226664 Free PMC article. - Molecular Architecture of the Essential Yeast Histone Acetyltransferase Complex NuA4 Redefines Its Multimodularity.
Setiaputra D, Ahmad S, Dalwadi U, Steunou AL, Lu S, Ross JD, Dong MQ, Côté J, Yip CK. Setiaputra D, et al. Mol Cell Biol. 2018 Apr 16;38(9):e00570-17. doi: 10.1128/MCB.00570-17. Print 2018 May 1. Mol Cell Biol. 2018. PMID: 29463645 Free PMC article. - N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex.
Wu WH, Wu CH, Ladurner A, Mizuguchi G, Wei D, Xiao H, Luk E, Ranjan A, Wu C. Wu WH, et al. J Biol Chem. 2009 Mar 6;284(10):6200-7. doi: 10.1074/jbc.M808830200. Epub 2008 Dec 16. J Biol Chem. 2009. PMID: 19088068 Free PMC article. - Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Qu K, Chen K, Wang H, Li X, Chen Z. Qu K, et al. Nature. 2022 Oct;610(7932):569-574. doi: 10.1038/s41586-022-05303-x. Epub 2022 Oct 5. Nature. 2022. PMID: 36198799 - Insights Into the Function of the NuA4 Complex in Plants.
Espinosa-Cores L, Bouza-Morcillo L, Barrero-Gil J, Jiménez-Suárez V, Lázaro A, Piqueras R, Jarillo JA, Piñeiro M. Espinosa-Cores L, et al. Front Plant Sci. 2020 Feb 21;11:125. doi: 10.3389/fpls.2020.00125. eCollection 2020. Front Plant Sci. 2020. PMID: 32153620 Free PMC article. Review.
References
- Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20:5695-5707. - PubMed
- Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321-329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials