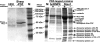Characterization of Staufen 1 ribonucleoprotein complexes - PubMed (original) (raw)
Characterization of Staufen 1 ribonucleoprotein complexes
Cornelia Brendel et al. Biochem J. 2004.
Abstract
In Drosophila oocytes and neuroblasts, the double-stranded RNA binding protein Staufen assembles into ribonucleoprotein particles, which mediate cytoplasmic mRNA trafficking and translation. Two different mammalian orthologues also appear to reside in distinct RNA-containing particles. To date, relatively little is known about the molecular composition of Staufen-containing ribonucleoprotein complexes. Here, we have used a novel one-step affinity purification protocol to identify components of Staufen 1-containing particles. Whereas the nucleocytoplasmic RNA-binding protein nucleolin is linked to Staufen in an RNA-dependent manner, the association of protein phosphatase 1, the microtubule-dependent motor protein kinesin and several components of the large and small ribosomal subunits with Staufen ribonucleoprotein complexes is RNA-independent. Notably, all these components do not co-purify with a second RNA-binding protein, hnRNPK (heterogeneous ribonucleoprotein K), demonstrating the high specificity of the purification protocol. Furthermore, pull-down and immunoprecipitation experiments suggest a direct interaction between Staufen 1 and the ribosomal protein P0 in vitro as well as in cells. In cell fractionation and sucrose gradient assays, Staufen co-fractionates with intact ribosomes and polysomes, but not with the isolated 40 S ribosomal subunit. Taken together, these findings imply that, in the cytoplasm of mammalian cells, an association with the ribosomal P-stalk protein P0 recruits Staufen 1 into ribosome-containing ribonucleoprotein particles, which also contain kinesin, protein phosphatase 1 and nucleolin.
Figures
Figure 1. One-step affinity purification of Stau1 ribonucleoprotein complexes
C-terminally tagged Sharpin–PDZ, hnRNPK–PDZ and Stau1–PDZ were affinity-purified with GKAP-coated Sepharose beads from untreated (−) and RNase-digested (+) extracts of transfected HEK-293 cells lysed in RIPA buffer. After washing, proteins were separated via gel electrophoresis. Prominent bands were excised (arrows) and proteins were identified by MS. Stau1 RNPs contain the RNA-binding proteins RNA helicase A, hnRNPU, NFAR and nucleolin, as well as several proteins from the large and small ribosomal subunit, all of which are not identified in precipitates obtained from untransfected (HEK), Sharpin–PDZ (Sharp-PDZ) or hnRNPK–PDZ-expressing cells. The increase in affinity-purification efficiency of tagged Stau1 after RNase treatment probably reflects an increased accessibility of PDZ domains in the complex. Incomplete Stau-PDZ: short variants of PDZ-tagged rStau1, which may result from partial degradation. M, marker lane.
Figure 2. Components of Stau1 RNPs
(A) Western blot analysis of Stau1–PDZ and hnRNPK–PDZ precipitates (P) and supernatant fractions (S) from untreated (−) and RNase-treated (+) HEK-293 cell lysates performed with the indicated antibodies. The ribosomal proteins P0 and L7a, as well as PP1 and kinesin, associate with Stau1 RNPs, but not hnRNPK complexes, in an RNA-independent manner. In contrast, the interaction of Stau1 with nucleolin and PABP is RNA-dependent. Whereas nucleolin is only found in the Stau1–PDZ pull-down, PABP is also detected in the hnRNPK–PDZ precipitate. In contrast with kinesin, the motor protein dynein is not found in the Stau1–PDZ pull-down. The prominent cytoplasmic protein EF1α is detected in none of the precipitates. (B) Both the 28 S and 18 S rRNAs are present in untreated (−), but not RNase-digested (+), lysates.
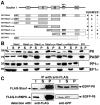
Figure 3. Mapping of the interaction domains in Stau1 and nucleolin
(A) Schematic representation of nucleolin, including the nuclear localization signal (NLS) as well as two different types of RNA-binding domains (RRM and RGG). Table 1 shows whether full-length Stau1 or its dsRBDs 2 and 3 alone co-immunoprecipitate with full-length nucleolin or C- and N-terminal portions of this protein. (B–E) Western blots of anti-FLAG immunoprecipitates obtained from HEK-293 cells co-expressing two different recombinant proteins (shown on the right). Fusion proteins were detected with either anti-FLAG or anti-GFP antibodies. I, input; S, supernatant fraction; P, pellet fraction. The interaction between Stau1 and nucleolin (Nuc) appears to involve RNA-binding domains in both proteins.
Figure 4. Stau1 co-fractionates with ribosomes
(A) Western blot of different rat brain fractions with anti-Stau1 serum. Lane 1, nuclear extract; lane 2, crude lysate; lane 3, cytosolic fraction; lane 4, interphase. Stau1 is highly enriched in the polysome fraction (lane 5). (B) Agarose gel analysis of total RNA isolated from Stau1–PDZ affinity-purified material reveals that it contains 18 S and 28 S rRNAs. (C) Analysis of 500 μl fractions from a 5–30% sucrose gradient performed with crude lysate from HEK-293 cells. Gel analysis of isolated RNA (uppermost row) and Western blot detection of P0, L7a, and Stau1 in individual fractions (lower three rows) is shown.
Figure 5. Interaction of Stau1 with ribosomal protein P0
(A) Schematic representation of Stau1, showing its dRBDs I–IV (boxes) and PP1 interaction domain (circle), as well as different GST fusion proteins. Table 1 summarizes whether the corresponding GST fusion proteins are able to pull-down rat brain P0, PP1 and PABP. (B) Western blots of GST pull-down experiments probed with specific antibodies against P0, PABP, PP1α and EF1α. Pull-downs were performed with the indicated GST fusion proteins and an adult rat brain lysate. P0 and PABP are only pulled-down by fusion proteins containing dsRBD III. PP1 precipitation requires the previously characterized PP1 interaction site. (C) Western blots of anti-FLAG immunoprecipitates obtained from HEK-293 cells co-expressing full-length or truncated versions of EGFP-tagged P0, together with either FLAG–Stau1 (upper row) or FLAG–hnRNPK (lower row). Recombinant proteins were detected with either anti-FLAG or anti-GFP antibodies. P0 co-immunoprecipitates with recombinant Stau1, but not with hnRNPK. I, input; S, supernatant fraction; P, pellet fraction.
Similar articles
- Barentsz, a new component of the Staufen-containing ribonucleoprotein particles in mammalian cells, interacts with Staufen in an RNA-dependent manner.
Macchi P, Kroening S, Palacios IM, Baldassa S, Grunewald B, Ambrosino C, Goetze B, Lupas A, St Johnston D, Kiebler M. Macchi P, et al. J Neurosci. 2003 Jul 2;23(13):5778-88. doi: 10.1523/JNEUROSCI.23-13-05778.2003. J Neurosci. 2003. PMID: 12843282 Free PMC article. - The RNA-binding protein Staufen from rat brain interacts with protein phosphatase-1.
Monshausen M, Rehbein M, Richter D, Kindler S. Monshausen M, et al. J Neurochem. 2002 May;81(3):557-64. doi: 10.1046/j.1471-4159.2002.00887.x. J Neurochem. 2002. PMID: 12065664 - Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes.
Thomas MG, Martinez Tosar LJ, Loschi M, Pasquini JM, Correale J, Kindler S, Boccaccio GL. Thomas MG, et al. Mol Biol Cell. 2005 Jan;16(1):405-20. doi: 10.1091/mbc.e04-06-0516. Epub 2004 Nov 3. Mol Biol Cell. 2005. PMID: 15525674 Free PMC article. - The role of mammalian Staufen on mRNA traffic: a view from its nucleocytoplasmic shuttling function.
Miki T, Takano K, Yoneda Y. Miki T, et al. Cell Struct Funct. 2005;30(2):51-6. doi: 10.1247/csf.30.51. Cell Struct Funct. 2005. PMID: 16377940 Review. - Staufen: from embryo polarity to cellular stress and neurodegeneration.
Tosar LJ, Thomas MG, Baez MV, Ibanez I, Chernomoretz A, Boccaccio GL. Tosar LJ, et al. Front Biosci (Schol Ed). 2012 Jan 1;4(2):432-52. doi: 10.2741/s277. Front Biosci (Schol Ed). 2012. PMID: 22202069 Review.
Cited by
- High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition.
Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC. Chiu YL, et al. Proc Natl Acad Sci U S A. 2006 Oct 17;103(42):15588-93. doi: 10.1073/pnas.0604524103. Epub 2006 Oct 9. Proc Natl Acad Sci U S A. 2006. PMID: 17030807 Free PMC article. - TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules.
Goulet I, Boisvenue S, Mokas S, Mazroui R, Côté J. Goulet I, et al. Hum Mol Genet. 2008 Oct 1;17(19):3055-74. doi: 10.1093/hmg/ddn203. Epub 2008 Jul 15. Hum Mol Genet. 2008. PMID: 18632687 Free PMC article. - The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization.
Chatel-Chaix L, Abrahamyan L, Fréchina C, Mouland AJ, DesGroseillers L. Chatel-Chaix L, et al. J Virol. 2007 Jun;81(12):6216-30. doi: 10.1128/JVI.00284-07. Epub 2007 Apr 11. J Virol. 2007. PMID: 17428849 Free PMC article. - Molecular composition of staufen2-containing ribonucleoproteins in embryonic rat brain.
Maher-Laporte M, Berthiaume F, Moreau M, Julien LA, Lapointe G, Mourez M, DesGroseillers L. Maher-Laporte M, et al. PLoS One. 2010 Jun 28;5(6):e11350. doi: 10.1371/journal.pone.0011350. PLoS One. 2010. PMID: 20596529 Free PMC article. - The double-stranded RNA-binding protein, Staufen1, is an IRES-transacting factor regulating HIV-1 cap-independent translation initiation.
Ramos H, Monette A, Niu M, Barrera A, López-Ulloa B, Fuentes Y, Guizar P, Pino K, DesGroseillers L, Mouland AJ, López-Lastra M. Ramos H, et al. Nucleic Acids Res. 2022 Jan 11;50(1):411-429. doi: 10.1093/nar/gkab1188. Nucleic Acids Res. 2022. PMID: 34893869 Free PMC article.
References
- Zhou Y., King M. L. Sending RNAs into the future: RNA localization and germ cell fate. IUBMB Life. 2004;56:19–27. - PubMed
- Jansen R. P. mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2001;2:247–256. - PubMed
- Li P., Yang X., Wasser M., Cai Y., Chia W. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell. 1997;90:437–447. - PubMed
- Buchner G., Bassi M. T., Andolfi G., Ballabio A., Franco B. Identification of a novel homolog of the drosophila staufen protein in the chromosome 8q13–q21.1 region. Genomics. 1999;62:113–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous