The DNA damage checkpoint pathways exert multiple controls on the efficiency and outcome of the repair of a double-stranded DNA gap - PubMed (original) (raw)
. 2004 Aug 10;32(14):4257-68.
doi: 10.1093/nar/gkh717. Print 2004.
Affiliations
- PMID: 15304563
- PMCID: PMC514360
- DOI: 10.1093/nar/gkh717
The DNA damage checkpoint pathways exert multiple controls on the efficiency and outcome of the repair of a double-stranded DNA gap
Edwin Haghnazari et al. Nucleic Acids Res. 2004.
Abstract
A DNA gap repair assay was used to determine the effect of mutations in the DNA damage checkpoint system on the efficiency and outcome (crossover/non-crossover) of recombinational DNA repair. In Saccharomyces cerevisiae gap repair is largely achieved by homologous recombination. As a result the plasmid either integrates into the chromosome (indicative of a crossover outcome) or remains extrachromosomal (indicative of a non-crossover outcome). Deletion mutants of the MEC1 and RAD53 checkpoint kinase genes exhibited a 5-fold decrease in gap repair efficiency, showing that 80% of the gap repair events depended on functional DNA damage checkpoints. Epistasis analysis suggests that the DNA damage checkpoints affect gap repair by modulating Rad51 protein-mediated homologous recombination. While in wild-type cells only approximately 25% of the gap repair events were associated with a crossover outcome, Mec1-deficient cells exhibited a >80% crossover association. Also mutations in the effector kinases Rad53, Chk1 and Dun1 were found to affect crossover association of DNA gap repair to various degrees. The data suggest that the DNA damage checkpoints are important for the optimal functioning of recombinational DNA repair with multiple terminal targets to modulate the efficiency and outcome of homologous recombination.
Figures
Figure 1
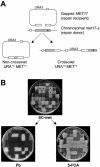
The double-stranded DNA gap repair system. (A) The ars plasmid containing the URA3 and MET17 markers is digested with BspEI and EcoNI to create a 238 bp gap leaving 1177 and 1021 bp of homology to the chromosomal met17-s gene on either sides of the gap, respectively. The chromosomal met17-s mutation is 216 bp downstream of the break site and is indicated as a black dot. The repaired plasmid can remain episomal (non-crossover) resulting in unstable transformants (MET+u URA+u) or integrate in the chromosome (crossover) resulting in stable transformants (MET+s URA+s). The figure is not drawn to scale. (B) Genetic analysis of the repair events. URA+ transformants are replica-plated onto SC-met media to determine, which transformants are also MET+. After several generations of non-selective growth, the transformants are replica-plated on 5-FOA plates and YPD plates containing lead nitrate (labeled Pb). Black dots represent URA+ MET+ transformants that are stable integrants indicative of crossover (MET+, white on lead plates; URA+, no growth on 5-FOA plates) and the white stars represent URA+ MET+ transformants that are unstable indicative of non-crossover events (MET−, black on lead plates; URA−, confluent growth on 5-FOA plates due to concomitant loss of both markers through plasmid loss under non-selective growth).
Figure 2
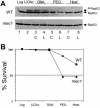
Transient activation of DNA damage checkpoints during transformation. (A) Phosphorylation status of Rad53 kinase was monitored at different stages of the transformation protocol by direct immunoblotting of whole cell extracts. Log = logarithmic cells washed once with 100 mM LiOAc solution; LiOAc = after 1 h incubation in 100 mM LiOAc; DNA = after 30 min incubation with either circular (C) or linear (L) transforming DNA; PEG = after 1 h incubation in PEG; Heat = after 15 min heat shock at 42°. WT, wild-type cells WDHY1800; mec1 WDHY1814. P-Rad53 indicates the phosphorylated form of Rad53 protein. (B) Survival of wild type and mec1 cells was determined at the identical time points as in (A). The averages of two experiments are presented.
Figure 3
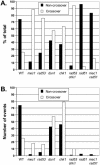
Distribution of crossover to non-crossover events. Graphic representation of the distribution of crossover (URA+s MET+s transformants) and non-crossover (URA+u MET+u transformants) events in wild-type and mutant S.cerevisiae strains. (A) Normalized data. The raw data are from Table 3 but were normalized to (URA+u MET+u events) + (URA+s MET+s events) as 100%. Except for the rad51 mec1 double mutant strain, these two classes represent 91% or more of the total URA+ MET+ transformants (see text). (B) Absolute data. The fractions of crossover and non-crossover events were multiplied by the gap repair efficiency (Table 2) and the data are plotted in comparison to 100 events in wild-type cells.
Figure 4
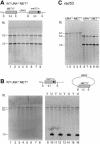
Physical analysis of transformants. (A) Total DNA isolated from eight YPD-grown URA+s MET+s wild-type transformants was doubly digested with BamHI (B) and SnaBI (S) and subjected to Southern analysis using a MET17 probe. The digest pattern indicates a simple integration of the plasmid as shown schematically above the Southern blot. (B) Total DNA isolated from eight YPD-grown URA+u MET+u wild-type transformants was doubly digested with BamHI (B) and SnaBI (S) and subjected to Southern analysis using a MET17 probe. The 6.7 kb band is indicative of the chromosomal met17-s as shown schematically above the Southern blots (lanes 1 to 8). When DNA was isolated from SC-ura-grown (U) cultures to select for the plasmid (lanes 10, 12, 14, 16), a prominent 1.5 kb band appeared, which represents the repaired MET17 ORF on the plasmid. The 1.5 kb band is absent in total DNA from YPD-grown (Y) transformants (lanes 9, 11, 13, 15), indicative of plasmid loss. The faint 5.7 kb band observed in SC-ura-grown lanes most likely represents non-specific hybridization of the probe with the abundant digested plasmid (7.2 kb plasmid − 1.5 kb MET17 ORF = 5.7 kb). (C) Total DNA isolated from rad53 transformants was doubly digested with BamHI and SnaBI and subjected to Southern analysis using a MET17 probe. All URA+s MET+s and URA+u MET+u transformants from each class exhibited the expected digest pattern.
Figure 5
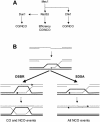
Model for checkpoint kinase-mediated control of DSB repair in S.cerevisiae. (A) DNA damage checkpoint pathways. Schematic representation of the organization of the Mec1, Rad53, Chk1 and Dun1 kinases in the DNA damage checkpoints in S.cerevisiae and the pathways to control efficiency and outcome of recombinational DNA repair. CO, crossover; NCO, non-crossover. For details see text. (B) Model for recombinational repair of DSBs. After DSB formation, the 5′ ends are resected to form 3′ ending single-stranded DNA tails. Rad51 protein-mediated DNA strand invasion leads to the formation of a D-loop. In DSBR, the second end of the DSB is captured by the D-loop leading to the formation of double Holliday junction, which can be resolved to crossover and non-crossover products. In SDSA, the invading strand reverses the D-loop and anneals with the second end to result in non-crossover products only. Dashed lines indicate newly synthesized DNA. CO: crossover, NCO: non-crossover. For details see text.
Similar articles
- RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates.
Bärtsch S, Kang LE, Symington LS. Bärtsch S, et al. Mol Cell Biol. 2000 Feb;20(4):1194-205. doi: 10.1128/MCB.20.4.1194-1205.2000. Mol Cell Biol. 2000. PMID: 10648605 Free PMC article. - Control of the yeast telomeric senescence survival pathways of recombination by the Mec1 and Mec3 DNA damage sensors and RPA.
Grandin N, Charbonneau M. Grandin N, et al. Nucleic Acids Res. 2007;35(3):822-38. doi: 10.1093/nar/gkl1081. Epub 2007 Jan 3. Nucleic Acids Res. 2007. PMID: 17202155 Free PMC article. - Meiotic roles of Mec1, a budding yeast homolog of mammalian ATR/ATM.
Carballo JA, Cha RS. Carballo JA, et al. Chromosome Res. 2007;15(5):539-50. doi: 10.1007/s10577-007-1145-y. Chromosome Res. 2007. PMID: 17674144 Review. - DNA structure dependent checkpoints as regulators of DNA repair.
Carr AM. Carr AM. DNA Repair (Amst). 2002 Dec 5;1(12):983-94. doi: 10.1016/s1568-7864(02)00165-9. DNA Repair (Amst). 2002. PMID: 12531008 Review.
Cited by
- Roles of exonucleases and translesion synthesis DNA polymerases during mitotic gap repair in yeast.
Guo X, Jinks-Robertson S. Guo X, et al. DNA Repair (Amst). 2013 Dec;12(12):1024-30. doi: 10.1016/j.dnarep.2013.10.001. Epub 2013 Nov 5. DNA Repair (Amst). 2013. PMID: 24210827 Free PMC article. - Combined Transcriptomics and Chemical-Genetics Reveal Molecular Mode of Action of Valproic acid, an Anticancer Molecule using Budding Yeast Model.
Golla U, Joseph D, Tomar RS. Golla U, et al. Sci Rep. 2016 Oct 13;6:35322. doi: 10.1038/srep35322. Sci Rep. 2016. PMID: 27734932 Free PMC article. - Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover.
Robert T, Dervins D, Fabre F, Gangloff S. Robert T, et al. EMBO J. 2006 Jun 21;25(12):2837-46. doi: 10.1038/sj.emboj.7601158. Epub 2006 May 25. EMBO J. 2006. PMID: 16724109 Free PMC article. - The extent of error-prone replication restart by homologous recombination is controlled by Exo1 and checkpoint proteins.
Tsang E, Miyabe I, Iraqui I, Zheng J, Lambert SA, Carr AM. Tsang E, et al. J Cell Sci. 2014 Jul 1;127(Pt 13):2983-94. doi: 10.1242/jcs.152678. Epub 2014 May 7. J Cell Sci. 2014. PMID: 24806966 Free PMC article. - Regulation of ribosomal DNA amplification by the TOR pathway.
Jack CV, Cruz C, Hull RM, Keller MA, Ralser M, Houseley J. Jack CV, et al. Proc Natl Acad Sci U S A. 2015 Aug 4;112(31):9674-9. doi: 10.1073/pnas.1505015112. Epub 2015 Jul 20. Proc Natl Acad Sci U S A. 2015. PMID: 26195783 Free PMC article.
References
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- Gerard F., Dri,A.M. and Moreau,P.L. (1999) Role of Escherichia coli RpoS, LexA and H-NS global regulators in metabolism and survival under aerobic, phosphate-starvation conditions. Microbiology, 145, 1547–1562. - PubMed
- Gellert M. (2002) V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem., 71, 101–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous