A CAF-1 dependent pool of HP1 during heterochromatin duplication - PubMed (original) (raw)
A CAF-1 dependent pool of HP1 during heterochromatin duplication
Jean-Pierre Quivy et al. EMBO J. 2004.
Abstract
To investigate how the complex organization of heterochromatin is reproduced at each replication cycle, we examined the fate of HP1-rich pericentric domains in mouse cells. We find that replication occurs mainly at the surface of these domains where both PCNA and chromatin assembly factor 1 (CAF-1) are located. Pulse-chase experiments combined with high-resolution analysis and 3D modeling show that within 90 min newly replicated DNA become internalized inside the domain. Remarkably, during this time period, a specific subset of HP1 molecules (alpha and gamma) coinciding with CAF-1 and replicative sites is resistant to RNase treatment. Furthermore, these replication-associated HP1 molecules are detected in Suv39 knockout cells, which otherwise lack stable HP1 staining at pericentric heterochromatin. This replicative pool of HP1 molecules disappears completely following p150CAF-1 siRNA treatment. We conclude that during replication, the interaction of HP1 with p150CAF-1 is essential to promote delivery of HP1 molecules to heterochromatic sites, where they are subsequently retained by further interactions with methylated H3-K9 and RNA.
Figures
Figure 1
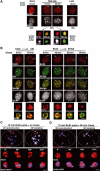
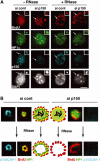
Specific dynamics of DNA synthesis in replicating pericentric heterochromatin. (A) Typical BrdU-labeled S-phase patterns. Top: 3T3 cells were pulsed for 10 or 90 min with BrdU (red, top) and stained with DAPI (bottom). Scale bar, 10 μm. Bottom: Magnification (six-fold) of individual replicating pericentric heterochromatin domain indicated by arrows on top. Corresponding BrdU (red), DAPI, and merge images are presented. In merge, DAPI was pseudocolored in green. Scale bar, 0.5 μm. (B) Newly replicated DNA is located at the periphery of the DAPI dense region and then becomes internalized. Left: Pulse–chase–pulse on 3T3 cells with first a CldU pulse followed by 45 min chase before a second IdU pulse was given, or both CldU and IdU were added together (0 min). Representative images (CldU, red; IdU, green) are shown with merge and DAPI staining. Right: 3T3 cells pulsed for 10 min with BrdU were chased for various times as indicated. Typical BrdU (red) and PCNA (green) patterns with merge and their corresponding DAPI staining are presented. Scale bar, 10 μm. The arrows point to the pericentric heterochromatin domain magnified six-fold below. Scale bar, 0.5 μm. (C) 3D reconstruction and modeling of DNA synthesis at pericentric heterochromatin domains. 3T3 cell were pulse labeled with BrdU for 10 min, and after immuno- and DAPI staining, z stack images of a single 3T3 cell nucleus in mid-late S-phase were used for 3D reconstruction and modeling. Localization of BrdU incorporation (red) and DNA (blue) in the whole nucleus. The dashed line represents the periphery of the nucleus. The arrow indicates the heterochromatin domain selected for magnification below. Scale bar, 10 μm. Several rotation angles are presented and ongoing DNA synthesis (red) is shown with DAPI staining (transparent blue). Scale bar, 0.5 μm. (D) As above, for 90 min chase.
Figure 2
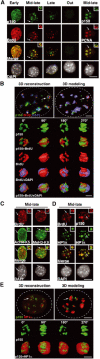
3D architecture of p150CAF-1, HP1, PCNA, and histones in replicating pericentric heterochromatin domains. (A) Mouse p150CAF-1 and DNA synthesis. p150CAF-1 (green) in combination with a 10 min pulse BrdU labeling (red, left), both during and outside S-phase, is shown in parallel with a double staining with PCNA (red, right) for mid-late S-phase only. Merge and DAPI images are presented. See typical foci in mid-late S-phase identified by an arrow at a three-fold magnification (insets). Scale bar, 10 μm. (B) 3D analysis of p150CAF-1 colocalizing with active DNA synthesis at the periphery of the pericentric heterochromatin domains. As in Figure 1C with p150CAF-1 (green), BrdU incorporation after a 10 min pulse (red), and DNA (blue). Scale bar, 1 μm. Bottom: The heterochromatin domain (see arrow above) is magnified four-fold and p150CAF-1 (green), BrdU (red), and DNA (blue) are shown either separately or merged for several rotation angles. The BrdU and DAPI stainings are shown transparently in the merge images. Scale bar, 0.5 μm. (C) In mid-late S-phase, AcH4-K5 and MeH3-K9 (green) were visualized in combination with BrdU incorporation (red) in mid-late S-phase. The arrow indicates typical foci magnified four-fold in the inset. Scale bar is as in (A). (D) Mouse p150CAF-1 and BrdU incorporation (red) were visualized in combination with HP1α (green) in mid-late S-phase. The arrow indicates typical foci magnified four-fold in the inset. Scale bar is as in (A). (E) 3D analysis as in (B) with p150CAF-1 (green) and HP1α (red and transparent).
Figure 3
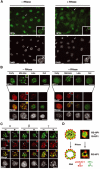
Replication-specific RNase-resistant HP1α and HP1γ but not HP1β pools around pericentric loci colocalizes with p150CAF-1 and BrdU. (A) A subset of cells retain some HP1α staining at pericentric heterochromatin domains following RNase treatment. A wide field showing HP1α labeling (green, top) and DAPI staining (bottom) in 3T3 cells without or with RNase treatment before fixation. The insets show higher magnifications focused on the cell indicated by the arrow as well as on individual foci magnified four-fold. Scale bar, 10 μm. (B) Only cells in mid-late S-phase retain significant HP1α staining as a rim around pericentromeric heterochromatin and sites of DNA synthesis. Double labeling in 3T3 cells of HP1α (green, top isolated) and BrdU (red) in 3T3 cells treated or not with RNase is shown for the early, mid-late, late, and out of S-phase. The arrow indicates typical foci magnified four-fold in the inset. The merge and corresponding DAPI staining are presented. Scale bar as in (A). (C) In mid-late S-phase cells, a fraction of HP1α and HP1γ but not HP1β is RNase-resistant and forms a rim around pericentromeric heterochromatin colocalizing with p150CAF-1. Double labeling of mid-late S-phase 3T3 cells with the three HP1 isoforms (green) and p150CAF-1 (red) without (−) or with (+) RNase treatment. Scale bar and arrows as in (B). (D) Model for HP1α and HP1γ distribution during replication of pericentromeric heterochromatin in mid-late S-phase. Top: Replicating pericentric heterochromatin is represented with a replication-independent pool of HP1 (RI-HP1, large light green circle) in the inner region interacting with RNA (black), and a replication-specific pool of HP1 at the periphery (RS-HP1, small dashed green circles). The location of p150CAF-1 or BrdU incorporation is shown (red circle). Colocalization of the DNA synthesis/p150CAF-1 site and RS-HP1 is represented in yellow. Bottom: Upon RNase treatment, the replication-independent pool of HP1 (RI-HP1) in the inner part is removed following RNA degradation without affecting the replication-specific pool of HP1 (RS-HP1) at the periphery. Corresponding stainings for p150CAF-1 (red) and HP1α (green) are shown as magnified images.
Figure 4
CAF-1 is found in HP1α/γ- and H3.1-containing complexes. (A) Analysis of e-HP1α-containing complexes. SDS–PAGE with fractions purified by FLAG and HA affinity (FLAG+HA), FLAG affinity (FLAG), and from glycerol gradient with the FLAG affinity-purified complex (FLAG glycerol gradient) stained with silver. The asterisk identifies the fraction used in (C). Positions corresponding to CAF-1 p150, p60, p48, HP1γ, and e-HP1α identified by mass spectrometry are indicated. Molecular weight markers (M) are shown. (B) Histones are not detected in the HP1α–CAF-1 complex. Western blot of complexes purified from mock, or e-HP1α- or e-H3.1-transduced HeLa cells. Proteins identified (right) and antibodies used (left) are indicated. (C) Comparison of the HP1α and H3.1 complexes in promoting nucleosome assembly coupled to DNA synthesis. Left: Supercoiling analysis. Cytosolic extract deficient in CAF-1 activity (p150) is used in combination with the HP1α–CAF-1 complex (fraction 5 of the glycerol gradient from (A), lane 1) and increasing amounts of the H3.1–CAF-1 complex (lanes 2–5). The migration of relaxed/nicked (Ir/II) and supercoiled circular DNA (I) is indicated. Right: Western blot analysis with samples corresponding to reactions on the left. Revelation with antibodies is as indicated.
Figure 5
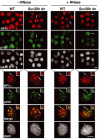
In Suv39h double mutant cells, a replication-specific pool of HP1α is colocalizing with p150CAF-1 at pericentric heterochromatin domains. Top: A wide field showing HP1α labeling (green), and p150CAF-1 (red) and DAPI staining in WT or Suv39h double-null (Suv39h dn) MEFs treated or not by RNase before fixation. The arrows indicate cells in mid-late S-phase based on p150CAF-1 staining. Scale bar, 10 μm. Bottom: As above with a mid-late S-phase nucleus. Merge and DAPI staining are presented. The arrow indicates typical foci magnified three-fold in the inset. Scale bar, 10 μm.
Figure 6
p150CAF-1 knock-down leads to a loss of HP1α at mid-late replication foci. (A) A concordant loss of p150CAF-1 at mid-late S-phase and the HP1 RNase-resistant fraction. Triple immunofluorescence labelings with BrdU (red, 10 min pulse), p150CAF-1 (green), and HP1α (blue) are shown for control or p150CAF-1 siRNA-treated 3T3 cells. Cells with mid-late S-phase profiles for various conditions are shown: nontreated (−RNase, left) or treated by RNase (+RNase, right). The arrow indicates a spot shown at a three-fold magnification in insets. Corresponding DAPI stainings are shown. Scale bar, 10 μm. (B) Model for the role of p150CAF-1 and HP1 localization at sites of DNA synthesis during replication of pericentromeric heterochromatin in mid-late S-phase. Left: In the presence of p150CAF-1 (si cont), replicating pericentric heterochromatin is represented with a replication-independent pool of HP1 (RI-HP1, large light green circle) in the inner region interacting with RNA (black), and a replication-specific pool of HP1 at the periphery (RS-HP1, small dashed green circles) colocalizing with p150CAF-1 (red circles). Colocalization of DNA synthesis and RS-HP1 is represented in yellow. Upon RNase treatment, the RI-HP1 in the inner part is removed following RNA degradation without affecting the RS-HP1 at the periphery. Right: In the absence of p150CAF-1 (si p150), the RS-HP1 (small dashed green circles) at the periphery is lost whereas the RI-HP1 (large light green circle) maintained by RNA (black) remains. Upon RNase treatment, all HP1 is thus lost. Corresponding magnified images from insets of (A) are shown. Stainings are p150CAF-1 (blue), and a merged image of HP1α (green) and BrdU (red).
Figure 7
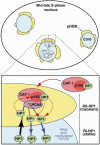
Hypothetical model for duplication of pericentric heterochromatin domains. Top: Scheme of a mouse cell nucleus in mid-late S-phase during replication of pericentric heterochromatin. A central core domain (blue) is surrounded by pHDB (yellow). Chromatin drawn in gray contains two forks emanating from one replication origin. Active synthesis takes place within this pHDB. Bottom: A model for a functional pHDB. For simplification, chromatin (gray) from only one fork is represented with one PCNA complex and no distinction is made between leading or lagging strand. The parental chromatin is pulled at the periphery (black arrow) to get replicated in pHDB (yellow). At the replication fork, PCNA (purple) targets p150CAF-1 (red) within the CAF-1 complex (pink), which could accept/transfer HP1 at the site of DNA synthesis and chromatin assembly (red dashed line). Constant HP1/chromatin ratio would be ensured by exchange with soluble CAF-1–HP1 complex (red solid arrow). Thereby a transient pool of replication-specific HP1 (RS-HP1, green, dashed line) localizes within pHDB. Synthesized and assembled daughter DNA is then pushed back inside the domain (blue arrows). The HP1 molecules from the parental chromatin can redistribute onto the newly replicated/assembled DNA (red dashed arrows) and would be stabilized in the inner domain as a stable pool of replication-independent HP1 (RI-HP1, green, solid line) by RNA and methylated H3-K9 as indicated.
Similar articles
- Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins.
Murzina N, Verreault A, Laue E, Stillman B. Murzina N, et al. Mol Cell. 1999 Oct;4(4):529-40. doi: 10.1016/s1097-2765(00)80204-x. Mol Cell. 1999. PMID: 10549285 - The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells.
Quivy JP, Gérard A, Cook AJ, Roche D, Almouzni G. Quivy JP, et al. Nat Struct Mol Biol. 2008 Sep;15(9):972-9. doi: 10.1038/nsmb.1470. Nat Struct Mol Biol. 2008. PMID: 19172751 - Pericentric heterochromatin generated by HP1 protein interaction-defective histone methyltransferase Suv39h1.
Muramatsu D, Singh PB, Kimura H, Tachibana M, Shinkai Y. Muramatsu D, et al. J Biol Chem. 2013 Aug 30;288(35):25285-25296. doi: 10.1074/jbc.M113.470724. Epub 2013 Jul 7. J Biol Chem. 2013. PMID: 23836914 Free PMC article. Retracted. - How HP1 Post-Translational Modifications Regulate Heterochromatin Formation and Maintenance.
Sales-Gil R, Vagnarelli P. Sales-Gil R, et al. Cells. 2020 Jun 12;9(6):1460. doi: 10.3390/cells9061460. Cells. 2020. PMID: 32545538 Free PMC article. Review. - Replication of heterochromatin: insights into mechanisms of epigenetic inheritance.
Wallace JA, Orr-Weaver TL. Wallace JA, et al. Chromosoma. 2005 Dec;114(6):389-402. doi: 10.1007/s00412-005-0024-6. Epub 2005 Nov 15. Chromosoma. 2005. PMID: 16220346 Review.
Cited by
- Mode of SUV420H2 heterochromatin localization through multiple HP1 binding motifs in the heterochromatic targeting module.
Nakao M, Sato Y, Aizawa A, Kimura H. Nakao M, et al. Genes Cells. 2024 May;29(5):361-379. doi: 10.1111/gtc.13109. Epub 2024 Feb 25. Genes Cells. 2024. PMID: 38403935 Free PMC article. - Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells.
Takami Y, Ono T, Fukagawa T, Shibahara K, Nakayama T. Takami Y, et al. Mol Biol Cell. 2007 Jan;18(1):129-41. doi: 10.1091/mbc.e06-05-0426. Epub 2006 Oct 25. Mol Biol Cell. 2007. PMID: 17065558 Free PMC article. - INCURVATA2 encodes the catalytic subunit of DNA Polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana.
Barrero JM, González-Bayón R, del Pozo JC, Ponce MR, Micol JL. Barrero JM, et al. Plant Cell. 2007 Sep;19(9):2822-38. doi: 10.1105/tpc.107.054130. Epub 2007 Sep 14. Plant Cell. 2007. PMID: 17873092 Free PMC article. - The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin.
Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A, Nakatani Y, Dent SY, Almouzni G. Loyola A, et al. EMBO Rep. 2009 Jul;10(7):769-75. doi: 10.1038/embor.2009.90. Epub 2009 Jun 5. EMBO Rep. 2009. PMID: 19498464 Free PMC article. - A separable domain of the p150 subunit of human chromatin assembly factor-1 promotes protein and chromosome associations with nucleoli.
Smith CL, Matheson TD, Trombly DJ, Sun X, Campeau E, Han X, Yates JR 3rd, Kaufman PD. Smith CL, et al. Mol Biol Cell. 2014 Sep 15;25(18):2866-81. doi: 10.1091/mbc.E14-05-1029. Epub 2014 Jul 23. Mol Biol Cell. 2014. PMID: 25057015 Free PMC article.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, Reuter G, Jenuwein T (1999) Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J 18: 1923–1938 - PMC - PubMed
- Aten JA, Bakker PJ, Stap J, Boschman GA, Veenhof CH (1992) DNA double labelling with IdUrd and CldUrd for spatial and temporal analysis of cell proliferation and DNA replication. Histochem J 24: 251–259 - PubMed
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 - PubMed
- Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T (2003) Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299: 721–725 - PubMed
- Cook PR (1999) The organization of replication and transcription. Science 284: 1790–1795 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous