The N-terminal region of the murine coronavirus spike glycoprotein is associated with the extended host range of viruses from persistently infected murine cells - PubMed (original) (raw)
The N-terminal region of the murine coronavirus spike glycoprotein is associated with the extended host range of viruses from persistently infected murine cells
Jeanne H Schickli et al. J Virol. 2004 Sep.
Abstract
Although murine coronaviruses naturally infect only mice, several virus variants derived from persistently infected murine cell cultures have an extended host range. The mouse hepatitis virus (MHV) variant MHV/BHK can infect hamster, rat, cat, dog, monkey, and human cell lines but not the swine testis (ST) porcine cell line (J. H. Schickli, B. D. Zelus, D. E. Wentworth, S. G. Sawicki, and K. V. Holmes, J. Virol. 71:9499-9507, 1997). The spike (S) gene of MHV/BHK had 63 point mutations and a 21-bp insert that encoded 56 amino acid substitutions and a 7-amino-acid insert compared to the parental MHV strain A59. Recombinant viruses between MHV-A59 and MHV/BHK were selected in hamster cells. All of the recombinants retained 21 amino acid substitutions and a 7-amino-acid insert found in the N-terminal region of S of MHV/BHK, suggesting that these residues were responsible for the extended host range of MHV/BHK. Flow cytometry showed that MHV-A59 bound only to cells that expressed the murine glycoprotein receptor CEACAM1a. In contrast, MHV/BHK and a recombinant virus, k6c, with the 21 amino acid substitutions and 7-amino-acid insert in S bound to hamster (BHK) and ST cells as well as murine cells. Thus, 21 amino acid substitutions and a 7-amino-acid insert in the N-terminal region of the S glycoprotein of MHV/BHK confer the ability to bind and in some cases infect cells of nonmurine species.
Figures
FIG. 1.
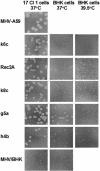
MHV/pi23 inefficiently infects hamster BHK cells. 17 Cl 1 cells and BHK cells were inoculated with MHV/pi23 at a multiplicity of infection of 3 to 5 (as determined by plaque assay in 17 Cl 1 cells) and fixed at 24 h postinoculation Viral nucleocapsid protein was detected by immunolabeling with anti-N MAb followed by fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin antibody. Magnification, 300×.
FIG. 2.
Comparison of the 12-bp insert of MHV/pi23 with the 21-bp insert of MHV/BHK. Compared to MHV-A59, MHV/pi23 has a 12-bp insert (bold). Eight of the nine nucleotides immediately upstream of the 12-bp inset of MHV/pi23 (italicized) match the 5′-most nine nucleotides (italicized) of the 21-bp insert (bold) of MHV/BHK.
FIG. 3.
Plaque morphology differs between recombinant viruses. Twice-plaque-purified recombinant viruses were plaqued in 17 Cl 1 cells at 37°C and BHK cells at 37 or 39.5°C. Plaques were visualized by neutral red staining and photographed 4 days postinoculation.
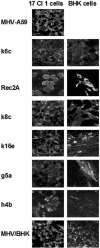
FIG. 4.
Expression of viral nucleocapsid protein differs between recombinant viruses. Following plaque purification, recombinant viruses were inoculated onto 17 Cl 1 or BHK cells at a multiplicity of infection of 3 to 5 PFU/cell, and the cells were fixed 24 h postinoculation. Viral nucleocapsid protein was visualized by labeling with anti-N MAb followed by fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin antibody. Magnification, 300×.
FIG. 5.
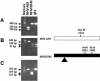
Restriction enzyme digestion analysis of recombinant viruses with HaeIII, AvrII, and NcoI. RT-PCR of the S genes of recombinant viruses was performed with primers 8 and 19 (Table 1). The RT-PCR products were digested with either HaeIII (A) or AvrII (B). In addition, RT-PCR of the S genes was performed with primers 9 and 16 (Table 1), and the RT-PCR products were digested by NcoI (C). MHV-A59 has one HaeIII site that yielded two fragments of 256 and 190 bp. MHV/BHK has one AvrII site that yielded two fragments of 349 and 97 bp and one NcoI site that yielded two fragments of 391 and 161 bp.
FIG. 6.
Recombinant viruses that infect BHK cells have the 5′ region of the S gene from MHV/BHK. Recombinant viruses had a single crossover event in one of three regions of the S gene, as determined by restriction enzyme digestion analysis and sequencing of RT-PCR products. Each of the representative recombinants shown retained the 21-bp insert (triangles) and 24 point mutations upstream of the insert found in the S gene of MHV/BHK.
FIG. 7.
Binding of viruses to cells of different species. Expression of mCEACAM1a on cells treated with the blocking anti-mCEACAM1a MAb CC1 or an isotype control MAb followed by phycoerythrin-conjugated goat anti-mouse immunoglobulin antibody is illustrated in panels A, B, C, D, and E. The percentage of cells positive for mCEACAM1a expression compared to the isotype control is also indicated. Binding of medium alone (no virus), MHV-A59, MHV/BHK, or the recombinant virus k6c to cells was detected with anti-S2 MAb 5B19 or an isotype control MAb followed by phycoerythrin-conjugated goat anti-mouse immunoglobulin antibody (F through Y). The percentage of cells positive for bound virus compared to the isotype control is also indicated. Flow cytometry data shown are representative of two or more independent experiments.
Similar articles
- The murine coronavirus mouse hepatitis virus strain A59 from persistently infected murine cells exhibits an extended host range.
Schickli JH, Zelus BD, Wentworth DE, Sawicki SG, Holmes KV. Schickli JH, et al. J Virol. 1997 Dec;71(12):9499-507. doi: 10.1128/JVI.71.12.9499-9507.1997. J Virol. 1997. PMID: 9371612 Free PMC article. - Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range.
de Haan CA, Te Lintelo E, Li Z, Raaben M, Wurdinger T, Bosch BJ, Rottier PJ. de Haan CA, et al. J Virol. 2006 Nov;80(22):10909-18. doi: 10.1128/JVI.00950-06. Epub 2006 Sep 6. J Virol. 2006. PMID: 16956938 Free PMC article. - The N-terminal domain of the murine coronavirus spike glycoprotein determines the CEACAM1 receptor specificity of the virus strain.
Tsai JC, Zelus BD, Holmes KV, Weiss SR. Tsai JC, et al. J Virol. 2003 Jan;77(2):841-50. doi: 10.1128/jvi.77.2.841-850.2003. J Virol. 2003. PMID: 12502800 Free PMC article. - Receptor specificity and receptor-induced conformational changes in mouse hepatitis virus spike glycoprotein.
Holmes KV, Zelus BD, Schickli JH, Weiss SR. Holmes KV, et al. Adv Exp Med Biol. 2001;494:173-81. doi: 10.1007/978-1-4615-1325-4_29. Adv Exp Med Biol. 2001. PMID: 11774465 Review. No abstract available. - Site directed mutagenesis of the murine coronavirus spike protein. Effects on fusion.
Bos EC, Heijnen L, Spaan WJ. Bos EC, et al. Adv Exp Med Biol. 1995;380:283-6. doi: 10.1007/978-1-4615-1899-0_45. Adv Exp Med Biol. 1995. PMID: 8830493 Review.
Cited by
- Three Amino Acid Substitutions in the Spike Protein Enable the Coronavirus Porcine Epidemic Diarrhea Virus To Infect Vero Cells.
Chen B, Dong S, Yu L, Si F, Li C, Xie C, Yu R, Li Z. Chen B, et al. Microbiol Spectr. 2023 Feb 14;11(1):e0387222. doi: 10.1128/spectrum.03872-22. Epub 2022 Dec 13. Microbiol Spectr. 2023. PMID: 36511700 Free PMC article. - Kathryn V. Holmes: A Career of Contributions to the Coronavirus Field.
Bonavia A, Dominguez SR, Dveksler G, Gagneten S, Howard M, Jeffers S, Qian Z, Smith MK, Thackray LB, Tresnan DB, Wentworth DE, Wessner DR, Williams RK, Miura TA. Bonavia A, et al. Viruses. 2022 Jul 20;14(7):1573. doi: 10.3390/v14071573. Viruses. 2022. PMID: 35891553 Free PMC article. Review. - Known Cellular and Receptor Interactions of Animal and Human Coronaviruses: A Review.
Everest H, Stevenson-Leggett P, Bailey D, Bickerton E, Keep S. Everest H, et al. Viruses. 2022 Feb 8;14(2):351. doi: 10.3390/v14020351. Viruses. 2022. PMID: 35215937 Free PMC article. Review. - Underlying selection for the diversity of spike protein sequences of SARS-CoV-2.
Ghosh M, Basak S, Dutta S. Ghosh M, et al. IUBMB Life. 2022 Mar;74(3):213-220. doi: 10.1002/iub.2577. Epub 2021 Nov 25. IUBMB Life. 2022. PMID: 34780121 Free PMC article. - Natural and Recombinant SARS-CoV-2 Isolates Rapidly Evolve In Vitro to Higher Infectivity through More Efficient Binding to Heparan Sulfate and Reduced S1/S2 Cleavage.
Shiliaev N, Lukash T, Palchevska O, Crossman DK, Green TJ, Crowley MR, Frolova EI, Frolov I. Shiliaev N, et al. J Virol. 2021 Oct 13;95(21):e0135721. doi: 10.1128/JVI.01357-21. Epub 2021 Aug 18. J Virol. 2021. PMID: 34406867 Free PMC article.
References
- Beauchemin, N., T. Chen, P. Draber, G. Dveksler, P. Gold, S. Gray-Owen, F. Grunert, S. Hammarstrom, K. V. Holmes, A. Karlson, M. Kuroki, S. H. Lin, L. Lucka, S. M. Najjar, M. Neumaier, B. Obrink, J. E. Shively, K. M. Skubitz, C. P. Stanners, P. Thomas, J. A. Thompson, M. Virji, S. von Kleist, C. Wagener, S. Watt, and W. Zimmermann. 1999. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp. Cell Res. 252:243-249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA046934/CA/NCI NIH HHS/United States
- R01 AI025231/AI/NIAID NIH HHS/United States
- R01-AI-25231/AI/NIAID NIH HHS/United States
- R01-AI-28506/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous