Lack of phylogeography in European mammals before the last glaciation - PubMed (original) (raw)
Lack of phylogeography in European mammals before the last glaciation
Michael Hofreiter et al. Proc Natl Acad Sci U S A. 2004.
Abstract
In many extant animal and plant species in Europe and North America a correlation exists between the geographical location of individuals and the genetic relatedness of the mitochondrial (mt) DNA sequences that they carry. Here, we analyze mtDNA sequences from cave bears, brown bears, cave hyenas, and Neandertals in Europe before the last glacial maximum and fail to detect any phylogeographic patterns similar to those observed in extant species. We suggest that at the beginning of the last glacial maximum, little phylogeographic patterns existed in European mammals over most of their geographical ranges and that current phylogeographic patterns are transient relics of the last glaciation. Cycles of retreat of species in refugia during glacial periods followed by incomplete dispersal from one refugium into other refugia during interglacial periods is likely to be responsible for the deep genetic divergences between phylogeographic clusters of mtDNA seen today.
Copyright 2004 The National Academy of Sciencs of the USA
Figures
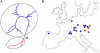
Fig. 1.
Phylogeography of Pleistocene cave bears. (A) Unrooted neighbor-joining tree for 26 cave bear mtDNA haplotypes. The longest branch divides the sequences into two clades present in 94% of bootstrap replicates. (B) Map of Europe showing the geographical distribution of the two cave bear mtDNA clades (in blue and red, respectively) before the last glacial maximum. Some dots represent more than one cave.
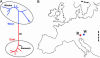
Fig. 2.
Phylogeography of Pleistocene brown bears. (A) Unrooted neighbor-joining tree for 18 modern and 2 ancient brown bear mtDNA haplotypes from Eurasia. The longest branch divides the sequences into two clades supported in 99% of bootstrap replicates. The “western” clade (blue) occurs today in western Europe and the “eastern” clade (red) in eastern Europe, Asia, and North America. The two Pleistocene mtDNA sequences are black. (B) Map of Europe showing the geographical location of the Pleistocene brown bears. R, Ramesch; W; Winden. The current areas of overlap between the two clades in Romania and Sweden are shown in black.
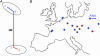
Fig. 3.
Phylogeography of Pleistocene cave hyenas. (A) Unrooted neighbor-joining tree for four cave hyena mtDNA haplotypes. The longest branch divides the sequences into two clades supported in 100% of bootstrap replicates. (B) Map of Europe showing the distribution of cave hyena mtDNA clades (blue and red, respectively) before the last glacial maximum.
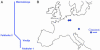
Fig. 4.
Neandertal mtDNA variation. (A) Unrooted neighbor-joining tree for four Neandertal mtDNA sequences. (B) Map showing the geographical provenience of the Neandertal individuals.
Fig. 5.
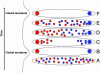
Schematic figure showing the possible effects of glacial cycles on phylogeographic mtDNA patterns. (A) A population that lacks phylogeographic structure is shown shortly before a glacial maximum. (B) During a glacial maximum only individuals in the refugia survive. By drift, different mtDNA types become fixed in the two refugia. (C) After the glaciation, recolonization from the refugia occurs. (D) Individuals from the two clades meet to form a “hybrid zone.” (E) Migration eventually erases the phylogeographic pattern for most of the population's range, but the higher population density slows migration into the refugia. (F) During a subsequent glacial maximum the refugia are likely to remain distinct with respect to mtDNA clades.
Similar articles
- Staying out in the cold: glacial refugia and mitochondrial DNA phylogeography in ancient European brown bears.
Valdiosera CE, García N, Anderung C, Dalén L, Crégut-Bonnoure E, Kahlke RD, Stiller M, Brandström M, Thomas MG, Arsuaga JL, Götherström A, Barnes I. Valdiosera CE, et al. Mol Ecol. 2007 Dec;16(24):5140-8. doi: 10.1111/j.1365-294X.2007.03590.x. Epub 2007 Nov 21. Mol Ecol. 2007. PMID: 18031475 - First DNA sequences from Asian cave bear fossils reveal deep divergences and complex phylogeographic patterns.
Knapp M, Rohland N, Weinstock J, Baryshnikov G, Sher A, Nagel D, Rabeder G, Pinhasi R, Schmidt HA, Hofreiter M. Knapp M, et al. Mol Ecol. 2009 Mar;18(6):1225-38. doi: 10.1111/j.1365-294X.2009.04088.x. Epub 2008 Jan 11. Mol Ecol. 2009. PMID: 19226321 - Mitochondrial DNA polymorphism, phylogeography, and conservation genetics of the brown bear Ursus arctos in Europe.
Taberlet P, Bouvet J. Taberlet P, et al. Proc Biol Sci. 1994 Mar 22;255(1344):195-200. doi: 10.1098/rspb.1994.0028. Proc Biol Sci. 1994. PMID: 8022838 - Impacts of Quaternary glaciation, geological history and geography on animal species history in continental East Asia: A phylogeographic review.
Fu J, Wen L. Fu J, et al. Mol Ecol. 2023 Aug;32(16):4497-4514. doi: 10.1111/mec.17053. Epub 2023 Jun 18. Mol Ecol. 2023. PMID: 37332105 Review. - Phylogeography reveals the complex impact of the Last Glacial Maximum on New Zealand's terrestrial biota.
Marske KA, Boyer SL. Marske KA, et al. J R Soc N Z. 2022 May 26;54(1):8-29. doi: 10.1080/03036758.2022.2079682. eCollection 2024. J R Soc N Z. 2022. PMID: 39439472 Free PMC article. Review.
Cited by
- Population dynamic of the extinct European aurochs: genetic evidence of a north-south differentiation pattern and no evidence of post-glacial expansion.
Mona S, Catalano G, Lari M, Larson G, Boscato P, Casoli A, Sineo L, Di Patti C, Pecchioli E, Caramelli D, Bertorelle G. Mona S, et al. BMC Evol Biol. 2010 Mar 26;10:83. doi: 10.1186/1471-2148-10-83. BMC Evol Biol. 2010. PMID: 20346116 Free PMC article. - Isotopic evidence for omnivory among European cave bears: Late Pleistocene Ursus spelaeus from the Peştera cu Oase, Romania.
Richards MP, Pacher M, Stiller M, Quilès J, Hofreiter M, Constantin S, Zilhão J, Trinkaus E. Richards MP, et al. Proc Natl Acad Sci U S A. 2008 Jan 15;105(2):600-4. doi: 10.1073/pnas.0711063105. Epub 2008 Jan 10. Proc Natl Acad Sci U S A. 2008. PMID: 18187577 Free PMC article. - Escape to Alcatraz: evolutionary history of slender salamanders (Batrachoseps) on the islands of San Francisco Bay.
Martínez-Solano I, Lawson R. Martínez-Solano I, et al. BMC Evol Biol. 2009 Feb 11;9:38. doi: 10.1186/1471-2148-9-38. BMC Evol Biol. 2009. PMID: 19210775 Free PMC article. - Exploring the phylogeography and population dynamics of the giant deer (Megaloceros giganteus) using Late Quaternary mitogenomes.
Rey-Iglesia A, Lister AM, Campos PF, Brace S, Mattiangeli V, Daly KG, Teasdale MD, Bradley DG, Barnes I, Hansen AJ. Rey-Iglesia A, et al. Proc Biol Sci. 2021 May 12;288(1950):20201864. doi: 10.1098/rspb.2020.1864. Epub 2021 May 12. Proc Biol Sci. 2021. PMID: 33977786 Free PMC article. - Phylogeographic history of grey wolves in Europe.
Pilot M, Branicki W, Jedrzejewski W, Goszczyński J, Jedrzejewska B, Dykyy I, Shkvyrya M, Tsingarska E. Pilot M, et al. BMC Evol Biol. 2010 Apr 21;10:104. doi: 10.1186/1471-2148-10-104. BMC Evol Biol. 2010. PMID: 20409299 Free PMC article.
References
- Avise, J. C. (2000) Phylogeography (Harvard Univ. Press, Cambridge, MA).
- Remington, C. (1968) Evol. Biol., 321–428.
- Taberlet, P., Fumagalli, L., Wust-Saucy, A. G. & Cosson, J. F. (1998) Mol. Ecol. 7, 453–464. - PubMed
- Hewitt, G. (1996) Biol. J. Linn. Soc., 247–276.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources