tRNA dynamics on the ribosome during translation - PubMed (original) (raw)
tRNA dynamics on the ribosome during translation
Scott C Blanchard et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Using single-molecule fluorescence spectroscopy, time-resolved conformational changes between fluorescently labeled tRNA have been characterized within surface-immobilized ribosomes proceeding through a complete cycle of translation elongation. Fluorescence resonance energy transfer was used to observe aminoacyl-tRNA (aa-tRNA) stably accommodating into the aminoacyl site (A site) of the ribosome via a multistep, elongation factor-Tu dependent process. Subsequently, tRNA molecules, bound at the peptidyl site and A site, fluctuate between two configurations assigned as classical and hybrid states. The lifetime of classical and hybrid states, measured for complexes carrying aa-tRNA and peptidyl-tRNA at the A site, shows that peptide bond formation decreases the lifetime of the classical-state tRNA configuration by approximately 6-fold. These data suggest that the growing peptide chain plays a role in modulating fluctuations between hybrid and classical states. Single-molecule fluorescence resonance energy transfer was also used to observe aa-tRNA accommodation coupled with elongation factor G-mediated translocation. Dynamic rearrangements in tRNA configuration are also observed subsequent to the translocation reaction. This work underscores the importance of dynamics in ribosome function and demonstrates single-particle enzymology in a system of more than two components.
Copyright 2004 The National Academy of Sciencs of the USA
Figures
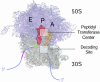
Fig. 1.
70S ribosome showing binding sites for A-, P-, and E-site tRNA molecules (model coordinates courtesy of M. Yusopov, Laboratoire de Biologie et Genomique Structurales de Institut de Génétique et de Biologie Moléculaire et Cellulaire, Strasbourg, France). The 50S subunit is shown in purple and the 30S subunit in tan. aa-tRNA (A), peptidyl-tRNA (P), and deacylated-tRNA (E) binding sites are labeled in black at their approximate location on the 70S ribosome. A-site tRNA is shown in gold and P-site tRNA in red; mRNA is shown in blue. The locations of the peptidyltransferase center and decoding site are highlighted.
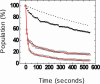
Fig. 2.
Single-molecule puromycin assay. Surface-immobilized ribosome complexes carrying Cy3-Met-tRNAfMet in the P site (dye-labeled at the α-amino group) react with puromycin. Stopped-flow addition of puromycin (1 mM, pH 7.5) to the immobilized complexes results in peptide bond formation and rapid loss of Cy3 signal caused by dissociation of the puromycin-Met-Cy3 product from the surface. Before A-site tRNA accommodation, the loss of fluorescence was fit to a double exponential, A_1*exp(–_t/τ1) + A_2* exp(–_t/τ2) + _y_0, where _A_1 and _A_2 represent the population (%) of fast and slow decay reactions (solid lines). The fitting parameters from a double-exponential fit were: _A_1 = 79, τ1 = 12.6 ± 0.3 s, _A_2 = 18, τ2 = 236 ± 26 s, and _y_0 = 3. After enzymatic delivery of Phe-tRNAPhe, the rate of puromycin reaction is dramatically reduced, consistent with tRNA accommodation at the A site and peptide bond formation. Less that 5% of the population shows a fast decay after Phe-tRNAPhe delivery. However, some residual reaction with puromycin remains. For comparison, an independent measurement of the intrinsic Cy3 signal decay caused by peptidyl-tRNA dissociation from the A site is shown (dotted line). Recovery of the rapid puromycin reaction was achieved by incubation with EF-G in the presence of GTP. The fitting parameters were _A_1 = 58, τ1 = 6.4 ± 0.3 s, _A_2 = 27, τ2 = 154 ± 8 s, and _y_0 = 15. Thus, ≈73% (58/79) of ribosomes return to reacting rapidly with puromycin.
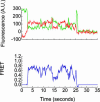
Fig. 3.
Single-molecule Cy3 and Cy5 fluorescence and FRET. tRNA delivery to ribosome complexes carrying fMet-tRNAfMet(Cy3-s4U) in the P site was monitored under Cy3 excitation. Cy5 fluorescence arises from Phe-tRNAPhe(Cy5-acp3U) binding to the ribosome and colocalization of Cy3 and Cy5 fluorophores within the same complex. (Upper) Cy3 and Cy5 emission intensity are shown in green and red, respectively. (Lower) The corresponding FRET value, _I_Cy5/(_I_Cy3 + _I_Cy5), is shown in blue.
Fig. 4.
Postsynchronized, time-resolved population FRET histograms reporting on intermolecular tRNA–tRNA distances within individual ribosomes of an ensemble of particles. Histograms show the evolution of FRET for an ensemble of surface-immobilized 70S complexes carrying tRNAfMet(Cy3-s4U) in the P site after delivery of EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U) where individual FRET time traces have been synchronized to the first FRET value ≥ 0.25. Population information is color-coded from tan (lowest occupancy) to red (highest occupancy). The histogram plots generated by stopped-flow delivery of EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U) depend on the acylation state of tRNA in the P site and in the presence of EF-G. (A) Complexes carrying fMet-tRNAfMet(Cy3-s4U) in the P site. (B) Complexes carrying OH-tRNAfMet(Cy3-s4U) in the P site. (C) Exactly as in A but in the presence of EF-G. Within experimental uncertainty, the high FRET peaks observed have the same mean FRET value, 0.74 ± 0.05. Similarly, the lower FRET peaks observed in A and B have a mean FRET value of 0.45 ± 0.05. Peaks centered at zero FRET arise from Cy5 blinking, photobleaching, and tRNA dissociation.
Similar articles
- Elongation factor-Tu can repetitively engage aminoacyl-tRNA within the ribosome during the proofreading stage of tRNA selection.
Morse JC, Girodat D, Burnett BJ, Holm M, Altman RB, Sanbonmatsu KY, Wieden HJ, Blanchard SC. Morse JC, et al. Proc Natl Acad Sci U S A. 2020 Feb 18;117(7):3610-3620. doi: 10.1073/pnas.1904469117. Epub 2020 Feb 5. Proc Natl Acad Sci U S A. 2020. PMID: 32024753 Free PMC article. - Dynamics of tRNA occupancy and dissociation during translation by the ribosome.
Xie P. Xie P. J Theor Biol. 2013 Jan 7;316:49-60. doi: 10.1016/j.jtbi.2012.09.023. Epub 2012 Sep 28. J Theor Biol. 2013. PMID: 23026766 - tRNA Fluctuations Observed on Stalled Ribosomes Are Suppressed during Ongoing Protein Synthesis.
Jamiolkowski RM, Chen C, Cooperman BS, Goldman YE. Jamiolkowski RM, et al. Biophys J. 2017 Dec 5;113(11):2326-2335. doi: 10.1016/j.bpj.2017.08.052. Biophys J. 2017. PMID: 29211986 Free PMC article. - The function of the translating ribosome: allosteric three-site model of elongation.
Rheinberger HJ. Rheinberger HJ. Biochimie. 1991 Jul-Aug;73(7-8):1067-88. doi: 10.1016/0300-9084(91)90149-u. Biochimie. 1991. PMID: 1742351 Review. - Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu.
Agirrezabala X, Frank J. Agirrezabala X, et al. Q Rev Biophys. 2009 Aug;42(3):159-200. doi: 10.1017/S0033583509990060. Q Rev Biophys. 2009. PMID: 20025795 Free PMC article. Review.
Cited by
- Transcription and Translation in Cytomimetic Protocells Perform Most Efficiently at Distinct Macromolecular Crowding Conditions.
Vibhute MA, Schaap MH, Maas RJM, Nelissen FHT, Spruijt E, Heus HA, Hansen MMK, Huck WTS. Vibhute MA, et al. ACS Synth Biol. 2020 Oct 16;9(10):2797-2807. doi: 10.1021/acssynbio.0c00330. Epub 2020 Oct 5. ACS Synth Biol. 2020. PMID: 32976714 Free PMC article. - Developing Multichannel smFRET Approach to Dissecting Ribosomal Mechanisms.
Lin R, Wang Y. Lin R, et al. Chem Biomed Imaging. 2024 Mar 21;2(7):501-509. doi: 10.1021/cbmi.4c00010. eCollection 2024 Jul 22. Chem Biomed Imaging. 2024. PMID: 39056063 Free PMC article. - Structural characterization of ribosome recruitment and translocation by type IV IRES.
Murray J, Savva CG, Shin BS, Dever TE, Ramakrishnan V, Fernández IS. Murray J, et al. Elife. 2016 May 9;5:e13567. doi: 10.7554/eLife.13567. Elife. 2016. PMID: 27159451 Free PMC article. - EF4 reveals the energy barrier for tRNA back-translocation in the peptidyl transferase center.
Song G, Qin Y. Song G, et al. RNA Biol. 2016 Oct 2;13(10):934-939. doi: 10.1080/15476286.2016.1215795. Epub 2016 Jul 29. RNA Biol. 2016. PMID: 27472653 Free PMC article. - Whither Ribosome Structure and Dynamics Research? (A Perspective).
Frank J. Frank J. J Mol Biol. 2016 Sep 11;428(18):3565-9. doi: 10.1016/j.jmb.2016.04.034. Epub 2016 May 10. J Mol Biol. 2016. PMID: 27178840 Free PMC article. Review.
References
- Wimberly, B. T., Brodersen, D. E., Clemons, W. M., Jr., Morgan-Warren, R. J., Carter, A. P., Vonrhein, C., Hartsch, T. & Ramakrishnan, V. (2000) Nature 407, 327–339. - PubMed
- Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F. & Yonath, A. (2000) Cell 102, 615–623. - PubMed
- Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000) Science 289, 905–920. - PubMed
- Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F. & Yonath, A. (2001) Cell 107, 679–688. - PubMed
- Yusupov, M. M., Yusupova, G. Z., Baucom, A., Lieberman, K., Earnest, T. N., Cate, J. H. & Noller, H. F. (2001) Science 292, 883–896. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources