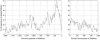Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification - PubMed (original) (raw)
Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification
Uwe Ohler et al. RNA. 2004 Sep.
Abstract
MicroRNAs are approximately 22-nucleotide (nt) RNAs processed from foldback segments of endogenous transcripts. Some are known to play important gene regulatory roles during animal and plant development by pairing to the messages of protein-coding genes to direct the post-transcriptional repression of these messages. Previously, we developed a computational method called MiRscan, which scores features related to the foldbacks, and used this algorithm to identify new miRNA genes in the nematode Caenorhabditis elegans. In the present study, to identify sequences that might be involved in processing or transcriptional regulation of miRNAs, we aligned sequences upstream and downstream of orthologous nematode miRNA foldbacks. These alignments showed a pronounced peak in sequence conservation about 200 bp upstream of the miRNA foldback and revealed a highly significant sequence motif, with consensus CTCCGCCC, that is present upstream of almost all independently transcribed nematode miRNA genes. Scoring the pattern of upstream/downstream conservation, the occurrence of this sequence motif, and orthology of host genes for intronic miRNA candidates, yielded substantial improvements in the accuracy of MiRscan. Nine new C. elegans miRNA gene candidates were validated using a PCR-sequencing protocol. As previously seen for bacterial RNA genes, sequence features outside of the RNA secondary structure can therefore be very useful for the computational identification of eukaryotic noncoding RNA genes. The total number of confidently identified nematode miRNAs now approaches 100. The improved analysis supports our previous assertion that miRNA gene identification is nearing completion in C. elegans with apparently no more than 20 miRNA genes now remaining to be identified.
Figures
FIGURE 1.
Conservation upstream and downstream of nematode microRNA foldbacks. The percentage of C. elegans sequences that are part of a conserved aligned block with C. briggsae at specific positions is plotted in bins of 10 bp. The positions are given relative to the beginning (left) or end (right) of the 110-nt segments containing the foldback. Genomic sequences were aligned using DBA and BayesBlockAligner as described in the text. Example alignments are part of the Supplementary Material (
http://genes.mit.edu/burgelab/MiRscanII
).
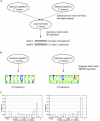
FIGURE 2.
Identification of conserved upstream sequence elements. (A) Enumerative search for over-represented 8-mers within conserved upstream regions. Next to each consensus sequence is the number of instances of this sequence in conserved C. elegans blocks allowing for zero or one mismatch to the consensus or its reverse complement, and the number of distinct upstream sequences containing these instances. The Z-score of the consensus motif A was 29.0, the score of motif B was 14.7. As a control, a search in equally sized, randomly generated sequences delivered a Z-score of 11.2. (B) Application of the MEME local alignment algorithm to the complete 2000-bp upstream sequence sets. Shown are the pictograms (
http://genes.mit.edu/pictogram.html
) computed from the sequences that were used in the alignment by MEME for C. elegans (E-value of 3.0e-24) and C. briggsae (E-value of 1.5e-37). Both methods identify a highly similar motif as the most significant one. (C) Histograms of the locations of the best hit per sequence to the motifs given in B, in bins of 100 bp.
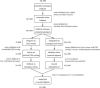
FIGURE 3.
Flowchart of filtering and rescoring of candidate foldbacks with MiRscanII. Input was the set of conserved foldbacks that had received scores by MiRscanI. The numbers show how many candidates passed each step.
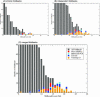
FIGURE 4.
Histograms of MiRscanII scores greater than zero (nonsyntenic analysis). (A) Intronic foldbacks. (B) Independent foldbacks. (C) Merged set of 13,398 foldbacks. The training set (orange), test set (dark blue), previously verified MiRscanI predictions (light blue), and newly verified MiRscanII predictions (red) are marked in color. The score distributions were truncated at 50 foldbacks on the y axis. The scores of one miRNA gene in the training set (mir-59) was negative, and thus is not shown. Each bin covers a score range of one bit, e.g., the bin labeled 15 includes candidates with scores between 15 and 16 bits.
Similar articles
- The microRNAs of Caenorhabditis elegans.
Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. Lim LP, et al. Genes Dev. 2003 Apr 15;17(8):991-1008. doi: 10.1101/gad.1074403. Epub 2003 Apr 2. Genes Dev. 2003. PMID: 12672692 Free PMC article. - Computational analysis of microRNA targets in Caenorhabditis elegans.
Watanabe Y, Yachie N, Numata K, Saito R, Kanai A, Tomita M. Watanabe Y, et al. Gene. 2006 Jan 3;365:2-10. doi: 10.1016/j.gene.2005.09.035. Epub 2005 Dec 13. Gene. 2006. PMID: 16356665 - Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans.
Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Ruby JG, et al. Cell. 2006 Dec 15;127(6):1193-207. doi: 10.1016/j.cell.2006.10.040. Cell. 2006. PMID: 17174894 - Functional genomic, computational and proteomic analysis of C. elegans microRNAs.
Lehrbach NJ, Miska EA. Lehrbach NJ, et al. Brief Funct Genomic Proteomic. 2008 May;7(3):228-35. doi: 10.1093/bfgp/eln024. Epub 2008 Jun 19. Brief Funct Genomic Proteomic. 2008. PMID: 18565984 Review. - Role of miRNA and miRNA processing factors in development and disease.
Conrad R, Barrier M, Ford LP. Conrad R, et al. Birth Defects Res C Embryo Today. 2006 Jun;78(2):107-17. doi: 10.1002/bdrc.20068. Birth Defects Res C Embryo Today. 2006. PMID: 16847880 Review.
Cited by
- Asymmetric purine-pyrimidine distribution in cellular small RNA population of papaya.
Aryal R, Yang X, Yu Q, Sunkar R, Li L, Ming R. Aryal R, et al. BMC Genomics. 2012 Dec 5;13:682. doi: 10.1186/1471-2164-13-682. BMC Genomics. 2012. PMID: 23216749 Free PMC article. - Advances in the techniques for the prediction of microRNA targets.
Zheng H, Fu R, Wang JT, Liu Q, Chen H, Jiang SW. Zheng H, et al. Int J Mol Sci. 2013 Apr 15;14(4):8179-87. doi: 10.3390/ijms14048179. Int J Mol Sci. 2013. PMID: 23591837 Free PMC article. Review. - Ab initio identification of human microRNAs based on structure motifs.
Brameier M, Wiuf C. Brameier M, et al. BMC Bioinformatics. 2007 Dec 18;8:478. doi: 10.1186/1471-2105-8-478. BMC Bioinformatics. 2007. PMID: 18088431 Free PMC article. - The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans.
Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. Abbott AL, et al. Dev Cell. 2005 Sep;9(3):403-14. doi: 10.1016/j.devcel.2005.07.009. Dev Cell. 2005. PMID: 16139228 Free PMC article. - miRRim: a novel system to find conserved miRNAs with high sensitivity and specificity.
Terai G, Komori T, Asai K, Kin T. Terai G, et al. RNA. 2007 Dec;13(12):2081-90. doi: 10.1261/rna.655107. Epub 2007 Oct 24. RNA. 2007. PMID: 17959929 Free PMC article.
References
- Abrahante, J.E., Daul, A.L., Li, M., Volk, L.M., Tennessen, J.M., Miller, E.A., and Rougvie, A.E. 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4: 625–637. - PubMed
- Ambros, V. 2003. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 113: 673–676. - PubMed
- Ambros, V., Lee, R.C., Lavanway, A., Williams, P.T., and Jewell, D. 2003b. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13: 807–818. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials